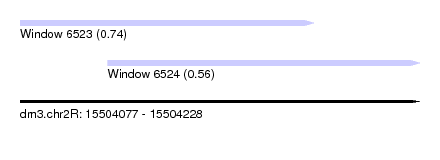
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,504,077 – 15,504,228 |
| Length | 151 |
| Max. P | 0.742616 |

| Location | 15,504,077 – 15,504,188 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.65 |
| Shannon entropy | 0.50382 |
| G+C content | 0.48965 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15504077 111 + 21146708 -------CCGAGCUUAAAUUGCAACCGCUGCAUUUGACGAUGCUCACGCUUUCAGUGGCUAACGCUGUACCCC--AACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUAUUGUCGU -------.(((........((((.....))))(((((.(((((....((((...(((((.((((.(.......--.).)))).)))))....))))....))))))))))......))). ( -27.80, z-score = -1.62, R) >droSim1.chr2R 14206664 100 + 19596830 -------CCGAGCUUAAAUUGCAACCGCUGCACUUA-----------GCUUCCAGUGGCUAACGCUGUUCCCC--UACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUACUGUCGU -------.(((((.......))....(.((((...(-----------((((...(((((.((((.((......--)).)))).)))))....)))))..)))).)...........))). ( -21.30, z-score = -0.87, R) >droSec1.super_1 13018672 100 + 14215200 -------CCGAGCUUAAAUUGCAACCGCUGCACUUA-----------GCUUCCAGUGGCUAACGCUGUUCCCC--UACCGUUCGCCGCAGUCAAGCUUCUGUAUCUCAAACUACUGUCGU -------..(((((.....((((.....))))...)-----------)))).(((((((.((((.((......--)).)))).)))((((........))))..........)))).... ( -21.10, z-score = -0.50, R) >droYak2.chr2R 7467554 103 - 21139217 CCUUAAAUGUGGCUUAAAUUGCAAGCGCUGCACUUGACGAUGCUCACGCUUGCAAUGGCUAACGAUAUUACCCCCAACCGUUUUUUACUGUGAAGCUUCUGUA----------------- .........(((((...((((((((((..(((........)))...)))))))))))))))..................(((((.......))))).......----------------- ( -23.40, z-score = -1.17, R) >droEre2.scaffold_4845 9701890 103 + 22589142 CCCUAAAUGGGGCUUAAACUGCAAGCGCUGCAUUUGACGAUGCUCACGCCUGCAAUGGCUAACGCUAUUCCUCAAUAGCGAUUUCCACAGUCGAGCUUCUGUA----------------- (((.....)))((((....((((.(((..(((((....)))))...))).)))).(((..(.(((((((....))))))).)..))).....)))).......----------------- ( -30.90, z-score = -1.79, R) >consensus _______CCGAGCUUAAAUUGCAACCGCUGCACUUGACGAUGCUCACGCUUCCAGUGGCUAACGCUGUUCCCC__UACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUA_UGUCGU .........((((((....((((.....))))......................(((((.((((..............)))).)))))....))))))...................... (-14.34 = -14.66 + 0.32)



| Location | 15,504,110 – 15,504,228 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.14 |
| Shannon entropy | 0.47087 |
| G+C content | 0.43276 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -10.66 |
| Energy contribution | -12.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15504110 118 + 21146708 UGCUCACGCUUUCAGUGGCUAACGCUGUACCCC--AACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUAUUGUCGUUCUGAUUGCCAUUUGGAUAUCUUUAGCUACUUAAAGAUAC ........((((.(((((((((....(.((..(--((..(((((..((((........))))....)))))..)))..)).).(((..((....))..))).))))))))).)))).... ( -24.60, z-score = -1.01, R) >droSim1.chr2R 14206693 111 + 19596830 -------GCUUCCAGUGGCUAACGCUGUUCCCC--UACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUACUGUCGUUCUGAUAAUCAUUUCGAUAUCUUUAGCUACUCAAAGAUAC -------.(((..(((((((((...........--....(((((..((((........))))....)))))...(((((...((.....))...)))))...)))))))))..))).... ( -22.90, z-score = -2.45, R) >droSec1.super_1 13018701 111 + 14215200 -------GCUUCCAGUGGCUAACGCUGUUCCCC--UACCGUUCGCCGCAGUCAAGCUUCUGUAUCUCAAACUACUGUCGUUCUGAUAAUCAUUUUGAUAUCUUUAUCUACUCAAAGAUAC -------((((...(((((.((((.((......--)).)))).)))))....))))....((((((........((((.....))))........((((....)))).......)))))) ( -20.60, z-score = -1.95, R) >droYak2.chr2R 7467594 101 - 21139217 UGCUCACGCUUGCAAUGGCUAACGAUAUUACCCCCAACCGUUUUUUACUGUGAAGCUUCUGUAUCU-----------------GAUAAUCAUUUGGAUAUCUUCAACUACCUCGAGAU-- ..(((..(((((((.(((..((((..............))))..))).))).))))....(((((.-----------------.(.......)..))))).............)))..-- ( -15.94, z-score = -0.45, R) >droEre2.scaffold_4845 9701930 102 + 22589142 UGCUCACGCCUGCAAUGGCUAACGCUAUUCCUCAAUAGCGAUUUCCACAGUCGAGCUUCUGUAUCU-----------------GAUAAUCACUGGGAUAUCUUAGGCUACGCAA-GAUAC .(((((((((......)))...(((((((....))))))).........)).))))....((((((-----------------........(((((....)))))........)-))))) ( -28.79, z-score = -2.21, R) >consensus UGCUCACGCUUCCAGUGGCUAACGCUGUUCCCC__UACCGUUUGCCACAGUCAAGCUUCUGUAUCUCAAACUA_UGUCGUUCUGAUAAUCAUUUGGAUAUCUUUAGCUACUCAAAGAUAC .......((((...(((((.((((..............)))).)))))....)))).........................................((((((((......)))))))). (-10.66 = -12.10 + 1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:18 2011