| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,473,940 – 15,474,035 |
| Length | 95 |
| Max. P | 0.920383 |

| Location | 15,473,940 – 15,474,035 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 60.10 |
| Shannon entropy | 0.81188 |
| G+C content | 0.52186 |
| Mean single sequence MFE | -16.54 |
| Consensus MFE | -6.80 |
| Energy contribution | -6.50 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15473940 95 + 21146708 --GUUGCAACAAUGCAACCCUCUCGGCCAUCGUC-CCCCACCCUUCCGAUUUUCCAUUUAAGCCCCACAUUCGCAUUGUUUUGCUGUGAUGCGUUCCA --((((((....))))))......((..((((..-...........))))...))......((..((((...(((......)))))))..))...... ( -16.72, z-score = -1.66, R) >droSim1.chr2R 14182925 95 + 19596830 --GUUGCAACAAUGCAACCCCCUCGCCACUCGCC-CCUCGCCCUUCCCAUUUUCCAUUUAAGCCCCACAUUCGCAUUGUUUUGCCGUGAUGCGUUCCA --((((((....)))))).....((((((..((.-....))...............................(((......))).)))..)))..... ( -13.20, z-score = -1.27, R) >droSec1.super_1 12993025 95 + 14215200 --GUUGCAACAAUGCAACCCCCUCACCACUCGCC-CCUCGCCCCUCCCAUUUUCCAUUUAAGCCCCACAUUCGCAUUGUUUUGCCGUGAUGCGUUCCA --((((((....))))))................-....................................(((((..(......)..)))))..... ( -13.10, z-score = -1.80, R) >droYak2.chr2R 7441132 87 - 21139217 --GUUGCAACAAUGCAACCCCCUCU---UUCGCC-CCG-----UCCCCAUUUUCCAUUUAAGCCCCACAUUCGCAUUGUUUUGCCGUGAUGCGUUCCA --((((((....)))))).......---......-...-----............................(((((..(......)..)))))..... ( -13.10, z-score = -1.97, R) >droEre2.scaffold_4845 9675784 96 + 22589142 --GUUGCAACAAUGCAGCCCCCCCGGCCACCACGGCCACCCCGUCCCCAUUUCCCAUUUAAGCCGUACAUUCGCAUUGUUUUGCCGUGCUGCGUUCCA --((((((....))))))......((((.....))))........................((.((((....(((......))).)))).))...... ( -22.60, z-score = -2.00, R) >droPer1.super_2 58645 87 - 9036312 AGGCUGCAGCA--GCUGCCACAUGGCUCAUGUGCACCCGCUCUCCGCUCAAUUCCAUCCGCAAAUC-CACUUGCAUUGUUCUACAA--AUGC------ .((.((((.((--...(((....)))...)))))).))((.....))............((((...-...))))............--....------ ( -18.40, z-score = 0.53, R) >dp4.chr3 12605588 87 + 19779522 AGGCUGCAGCA--GCUGCCACAUGGCUCAUGUGCACCCGCUCUCCGCUCAAUUCCAUCCGCAAAUC-CACUCGCAUUGUUCUACAA--AUGC------ .((.((((.((--...(((....)))...)))))).))((.....))...................-.....(((((........)--))))------ ( -18.50, z-score = 0.26, R) >droWil1.scaffold_180701 1508749 83 + 3904529 --GUUGCAUCG--GCAACCAAAC-----AUUUGUAUAUA----CAACAUACGUACAUUGUAAGGCA--AUUCCUAUUGUUAGUUUGGUGCACAUUCUA --.........--(((.((((((-----...((((..((----.....))..))))......((((--((....)))))).)))))))))........ ( -16.70, z-score = -0.68, R) >consensus __GUUGCAACAAUGCAACCCCCUCGCCCAUCGCC_CCCCCCCCCCCCCAUUUUCCAUUUAAGCCCCACAUUCGCAUUGUUUUGCCGUGAUGCGUUCCA ..(((((......)))))......................................................((((..(......)..))))...... ( -6.80 = -6.50 + -0.30)

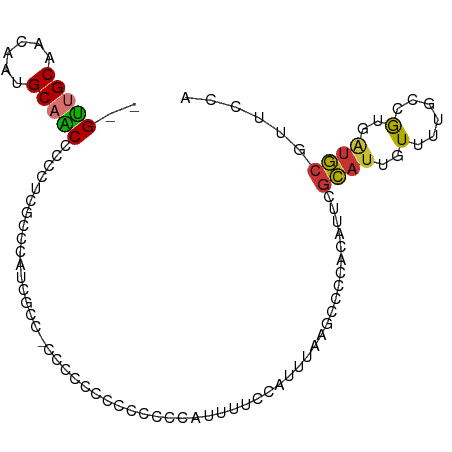
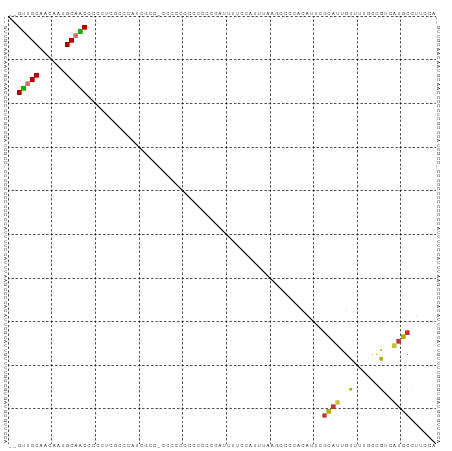
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:13 2011