
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,466,387 – 15,466,481 |
| Length | 94 |
| Max. P | 0.658204 |

| Location | 15,466,387 – 15,466,481 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 64.16 |
| Shannon entropy | 0.62623 |
| G+C content | 0.46354 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -10.53 |
| Energy contribution | -9.56 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
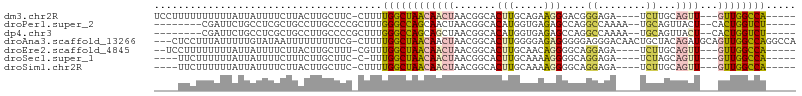
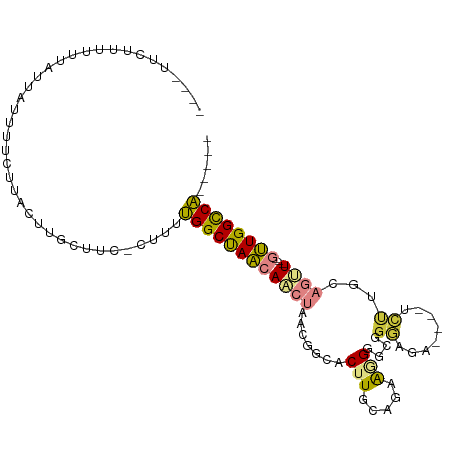
>dm3.chr2R 15466387 94 - 21146708 UCCUUUUUUUUUUAUUAUUUUCUUACUUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGCAGAAGGGACGGGAGA----UCUUGCAGUU---GUUGGCCA----- .................................-....((((((((((((....((....))....(((((....).----))))..))))---))))))))----- ( -21.20, z-score = -0.93, R) >droPer1.super_2 50630 90 + 9036312 --------CGAUUCUGCCUCGCUGCCUUGCCCCGCUUUGGGCCAGCAACUAACGGCACAUGGUGAGAGCCAGGCCAAAA--UGCAGUUACU--CACUGGUCU----- --------.((..((((...((((....((((......)))))))).......(((...((((....)))).)))....--.))))....)--)........----- ( -29.70, z-score = -0.39, R) >dp4.chr3 12597528 90 - 19779522 --------CGAUUCUGCCUCGCUGCCUUGCCCCGCUUUGGGCCAGCAGCUAACGGCACAUGGUGAGAGCCAGGCCAAAA--UGCAGUUACU--CACUGGUCU----- --------.((..((((...(((((...((((......))))..)))))....(((...((((....)))).)))....--.))))....)--)........----- ( -33.90, z-score = -1.10, R) >droAna3.scaffold_13266 10073285 103 + 19884421 ---CUCCUUUAUUUUUGUAUAAUUUUUUUUUCG-CUUUUGGCUAACAACUAACGGCACUUGGGGAGAGGGGGAGGGACAACUGCUACAGAUGCAGUUGGCCAGGCCA ---(((((((((......))))..........(-((.((((.......)))).))).....)))))....((..((.(((((((.......))))))).))...)). ( -27.10, z-score = -0.74, R) >droEre2.scaffold_4845 9665742 92 - 22589142 --UCCUUUUUUUUAUUAUUUUCUUACUUGCUUU-CGUUUGGCUAACAACUAACGGCACUUGCAACAGGGGCAGGAGA----UCUUGCAGUU---GUUGGCCA----- --...............................-....((((((((((((....((....)).......(((((...----.)))))))))---))))))))----- ( -25.60, z-score = -2.55, R) >droSec1.super_1 12985456 89 - 14215200 ----UUCUUUUUUAUUAUUUUCUUUCUUGCUUC-C-UUUGGCUAACAACUAACGGCACUUGCAAAAGGGGCAGGAGA----UCUAGCAGUU---GUUGGCCA----- ----.............................-.-..((((((((((((...(((.(((((.......))))).).----))....))))---))))))))----- ( -22.10, z-score = -1.04, R) >droSim1.chr2R 14167422 90 - 19596830 ----UUCUUUUUUAUUAUUUUCUUACUUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGCAAAAGGGGCAGGAGA----UCUUGCAGUU---GUUGGCCA----- ----.............................-....((((((((((((....((....)).......(((((...----.)))))))))---))))))))----- ( -25.60, z-score = -2.54, R) >consensus ____UUCUUUUUUAUUAUUUUCUUACUUGCUUC_CUUUUGGCUAACAACUAACGGCACUUGCAGAAGGGGCGGGAGA____UCUUGCAGUU___GUUGGCCA_____ ......................................((((((((..(....)...(((.....)))..........................))))))))..... (-10.53 = -9.56 + -0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:12 2011