| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,302,098 – 3,302,242 |
| Length | 144 |
| Max. P | 0.757158 |

| Location | 3,302,098 – 3,302,242 |
|---|---|
| Length | 144 |
| Sequences | 4 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.35379 |
| G+C content | 0.41429 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -19.93 |
| Energy contribution | -21.81 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3302098 144 + 23011544 ACAGGGGCGGACUU-AAAACCUAUUCAAGGGGGUUGGAGUUAAAUUUUGAAAUUAUUUGU----GUAUCUGAAGACAUGCGGGCAUAGUAAACACA----UACG----ACAAAUUACAGUAUCUACAUUUUCUC--AAAACCCCCUUUCUACGCCACUG .(((.(((((....-....)).....(((((((((.(((..((((...((.(((((((((----((((.((...(((((....))).))...)).)----))).----))))))...))).))...)))).)))--..))))))))).....))).))) ( -36.70, z-score = -2.08, R) >droYak2.chr2L 3286446 159 + 22324452 ACAGGGGCGUGCUCCAAAGCAAAUGCAAUGGGGUUGAAGUUGAAUUCCCAAGUUCUGUAUUCCGGUAUAGGAAGACAUGCUGGCAUACUAAGCACAUACAUACAUAUAACAAAUAACAGUCUGUGGUUUGCCCCCAAAAAUCCCCUCUCUGCGCCCCUG .((((((((((((....)))........(((((..(((.(((......))).))).((((.(((((((.(.....)))))))).))))...(((..((((.((...............)).))))...))))))))..............))))))))) ( -48.46, z-score = -1.95, R) >droSec1.super_5 1444535 147 + 5866729 ACAGGGGCGGAUUU-AAAAAAGAUUCAAGGGGGUUGAAGUUACAUGUUUAAGUUAUUUGU----GCAUCUGAAGACAAGCAGGCAUACUAAGCACAUAGAUACA----AUAAAUUACAGUAUCUGCGUUUCCUC--AAA-CCCCCUUUCUACGCCACUG .(((.(((((((((-.....))))))(((((((((((.(..((.((((((.((.....((----((..(((........)))))))))))))))..(((((((.----..........))))))).))..).))--.))-))))))).....))).))) ( -41.70, z-score = -2.91, R) >droSim1.chr2L 3239653 147 + 22036055 ACAGGGGCGGACUU-AAAACAGAUUCAAGGGGGUUUAAGUUACAUGUUUAAGUUAUUUGU----GUAUCUAAAGACAAGCGGGCAUAAUAAACACAUAGAUACA----AUAAAUUACAGUAUCUGCGCUACCUC--AAA-CCCCCUUUCUACGCCACUG .(((.((((((.((-......)).))((((((((((.((............(((..((((----((..((.......))...))))))..)))...(((((((.----..........)))))))......)).--)))-)))))))....)))).))) ( -36.50, z-score = -2.01, R) >consensus ACAGGGGCGGACUU_AAAACAGAUUCAAGGGGGUUGAAGUUAAAUGUUUAAGUUAUUUGU____GUAUCUGAAGACAAGCGGGCAUACUAAACACAUAGAUACA____ACAAAUUACAGUAUCUGCGUUUCCUC__AAA_CCCCCUUUCUACGCCACUG .(((.((((.................(((((((((................(((..((((....((((........))))....))))..)))...(((((((...............))))))).............)))))))))....)))).))) (-19.93 = -21.81 + 1.88)


| Location | 3,302,098 – 3,302,242 |
|---|---|
| Length | 144 |
| Sequences | 4 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.35379 |
| G+C content | 0.41429 |
| Mean single sequence MFE | -43.69 |
| Consensus MFE | -20.37 |
| Energy contribution | -21.81 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
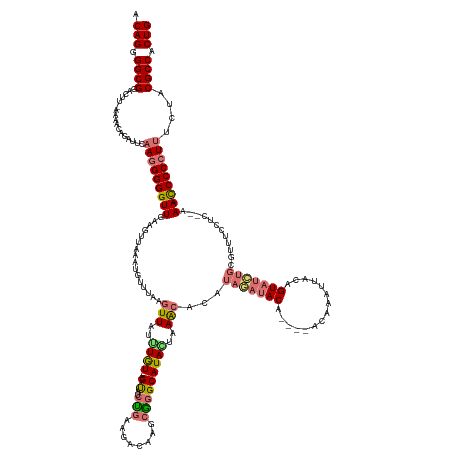
>dm3.chr2L 3302098 144 - 23011544 CAGUGGCGUAGAAAGGGGGUUUU--GAGAAAAUGUAGAUACUGUAAUUUGU----CGUA----UGUGUUUACUAUGCCCGCAUGUCUUCAGAUAC----ACAAAUAAUUUCAAAAUUUAACUCCAACCCCCUUGAAUAGGUUUU-AAGUCCGCCCCUGU (((.(((.....(((((((((..--(((.((((...((....((.((((((----.(((----(.((...((.(((....)))))...)).))))----)))))).)).))...))))..))).))))))))).....((....-....))))).))). ( -38.80, z-score = -2.54, R) >droYak2.chr2L 3286446 159 - 22324452 CAGGGGCGCAGAGAGGGGAUUUUUGGGGGCAAACCACAGACUGUUAUUUGUUAUAUGUAUGUAUGUGCUUAGUAUGCCAGCAUGUCUUCCUAUACCGGAAUACAGAACUUGGGAAUUCAACUUCAACCCCAUUGCAUUUGCUUUGGAGCACGCCCCUGU ((((((((....(((((((((((..((((((.((((((.((...(((.......)))...)).))))....)).))))....(((.((((......)))).)))...))..)))))))..))))....(((..((....))..)))....)))))))). ( -51.90, z-score = -1.43, R) >droSec1.super_5 1444535 147 - 5866729 CAGUGGCGUAGAAAGGGGG-UUU--GAGGAAACGCAGAUACUGUAAUUUAU----UGUAUCUAUGUGCUUAGUAUGCCUGCUUGUCUUCAGAUGC----ACAAAUAACUUAAACAUGUAACUUCAACCCCCUUGAAUCUUUUUU-AAAUCCGCCCCUGU (((.((((....(((((((-((.--((((..(((.((((((..........----.)))))).(((((..((((....)))).(((....)))))----))).............)))..)))))))))))))(((.....)))-.....)))).))). ( -40.60, z-score = -2.53, R) >droSim1.chr2L 3239653 147 - 22036055 CAGUGGCGUAGAAAGGGGG-UUU--GAGGUAGCGCAGAUACUGUAAUUUAU----UGUAUCUAUGUGUUUAUUAUGCCCGCUUGUCUUUAGAUAC----ACAAAUAACUUAAACAUGUAACUUAAACCCCCUUGAAUCUGUUUU-AAGUCCGCCCCUGU (((.(((((((((((((((-(((--(((........(((((..........----.)))))((((((((((...........((((....)))).----..........)))))))))).)))))))))))))...))))....-.....)))).))). ( -43.45, z-score = -3.20, R) >consensus CAGUGGCGUAGAAAGGGGG_UUU__GAGGAAACGCAGAUACUGUAAUUUAU____UGUAUCUAUGUGCUUAGUAUGCCCGCAUGUCUUCAGAUAC____ACAAAUAACUUAAAAAUGUAACUUCAACCCCCUUGAAUCUGUUUU_AAGUCCGCCCCUGU (((.((((....(((((((((....(((...(((.((((((...............)))))).)))................((((....))))..........................))).))))))))).................)))).))). (-20.37 = -21.81 + 1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:34 2011