
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,465,239 – 15,465,333 |
| Length | 94 |
| Max. P | 0.859810 |

| Location | 15,465,239 – 15,465,333 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 64.46 |
| Shannon entropy | 0.67238 |
| G+C content | 0.52653 |
| Mean single sequence MFE | -19.11 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
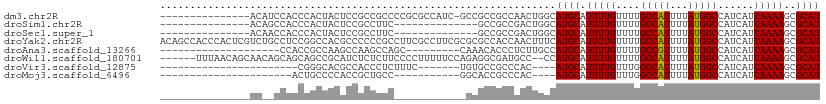
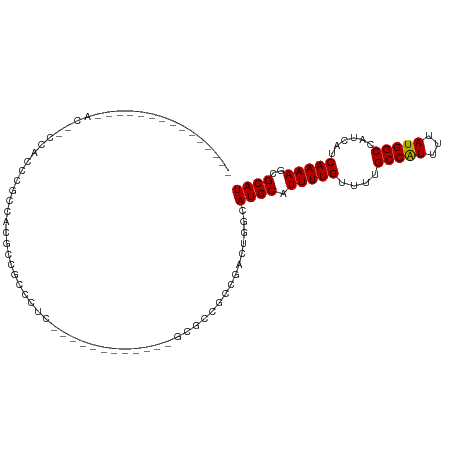
>dm3.chr2R 15465239 94 + 21146708 ---------------ACAUCCACCCACUACUCCGCCGCCCCGCGCCAUC-GCCGCCGCCAACUGGCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ---------------..........................((((....-...((.(((....)))..)).(((((....(((((...)))))......))))))))).. ( -20.00, z-score = -0.33, R) >droSim1.chr2R 14166337 81 + 19596830 ---------------ACAGCCACCCACUACUCCGCCUUC--------------GCCGCCGACUGGCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ---------------...((.............(((.((--------------(....)))..)))..((.(((((....(((((...)))))......))))))))).. ( -17.40, z-score = -0.75, R) >droSec1.super_1 12984379 81 + 14215200 ---------------ACAACCACCCACUACUCCGCCUUC--------------GCCGCCGACUGGCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ---------------..................(((.((--------------(....)))..)))((((.(((((....(((((...)))))......)))))..)))) ( -17.20, z-score = -1.29, R) >droYak2.chr2R 7431242 110 - 21139217 ACAGCCACCCACUCGUCUGCCUCCGGCCACGCCCCCCGCCUUCGCCUUCGCGCGCCACCAACUUUCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU .................(((....(((..(((.....((....))....))).)))............((.(((((....(((((...)))))......)))))))))). ( -19.40, z-score = 0.11, R) >droAna3.scaffold_13266 10072340 81 - 19884421 --------------------CCACCGCCAAGCCAAGCCAGC---------CAAACACCCUCUUGCCAUGCAUUUUGUUUUGCCGUUUUAUGGCCAUCAUCAAAAGCGCAU --------------------.....((((....((((..((---------.(((((......(((...)))...))))).)).))))..))))................. ( -10.90, z-score = 1.31, R) >droWil1.scaffold_180701 1494457 102 + 3904529 ------UUUAACAGCAACAGCAGCAGCCGCAUCUCUCUUCCCCUUUUUCCAGAGGCGAUGCC--CCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ------.......((....)).((....((((((((((............))))).))))).--....((.(((((....(((((...)))))......))))))))).. ( -22.90, z-score = -1.21, R) >droVir3.scaffold_12875 16366620 76 - 20611582 -----------------------CGGGCACGCCACCCUCUUUC-------UGUGCCGCCCAC----AUGCAUUUUGUUUGGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU -----------------------.((((..((((.........-------)).)).))))..----((((.(((((..(((((((...)))))))....)))))..)))) ( -25.20, z-score = -2.47, R) >droMoj3.scaffold_6496 20805570 73 + 26866924 ----------------------ACUGCCCCACCGCUGCC-----------GGCACCGCCCAC----AUGCAUUUUGUUUGGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ----------------------..(((......((((..-----------(((...)))..)----).)).(((((..(((((((...)))))))....)))))..))). ( -19.90, z-score = -0.71, R) >consensus _______________AC__CCACCCGCCACGCCGCCCUC____________GCGCCGCCGACUGGCAUGCAUUUUGUUUUGCCAUUUUAUGGCCAUCAUCAAAAGCGCAU ..................................................................((((.(((((....(((((...)))))......)))))..)))) (-10.68 = -10.58 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:11 2011