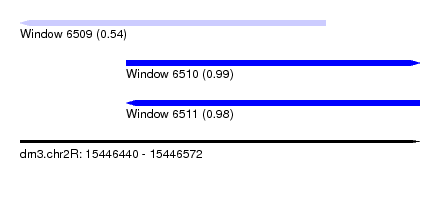
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,446,440 – 15,446,572 |
| Length | 132 |
| Max. P | 0.993391 |

| Location | 15,446,440 – 15,446,541 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.36140 |
| G+C content | 0.41154 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
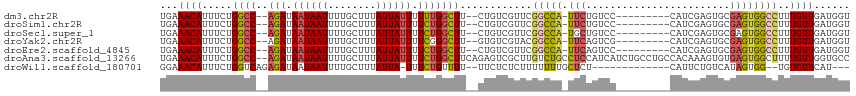
>dm3.chr2R 15446440 101 - 21146708 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUUUGGCUU--CUGUCGUUCGGCCA-UUCUGUCC---------CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ............((((--(((.((((((........)))))).))))))).--((((((...(((((-(((.(..(---------......)..))))))))).....)))))). ( -28.60, z-score = -2.29, R) >droSim1.chr2R 14147772 101 - 19596830 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUU--CUGUCGUUCGGCCA-UUCUGUCC---------CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ............((((--(((.((((((........)))))).))))))).--((((((...(((((-(((.(..(---------......)..))))))))).....)))))). ( -31.20, z-score = -3.09, R) >droSec1.super_1 12965880 101 - 14215200 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUU--CUGUCGUUCGGCCA-UGCUGUCC---------CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ............((((--(((.((((((........)))))).))))))).--((((((...(((((-(..((..(---------......)..)).)))))).....)))))). ( -28.90, z-score = -1.91, R) >droYak2.chr2R 7411369 101 + 21139217 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUCGGGCUU--GUGUCGUACGGCCA-UUCAGUCG---------CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ....((((....((((--.((.((((((........)))))).)).)))).--)))).....(((((-(((.(.((---------(.....))))))))))))............ ( -28.10, z-score = -1.07, R) >droEre2.scaffold_4845 9645808 101 - 22589142 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUU--CUGUCGUUCGGCCA-UUCAGUCC---------CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ............((((--(((.((((((........)))))).))))))).--((((((...(((((-(((.(..(---------......)..))))))))).....)))))). ( -31.20, z-score = -2.87, R) >droAna3.scaffold_13266 10053266 113 + 19884421 UGAAACAUUUCUGGCC--AGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCAGAGUCGCUUGUCUGCCUCCAUCAUCUGCCUGCCACAAAGUGUGAGUGGCUUUUGUUGGUGCC ........((((((((--(((.((((((........)))))).))))))..)))))..........((..(((.((...(((.(((((.....))).)))))...)).))).)). ( -32.00, z-score = -1.94, R) >droWil1.scaffold_180701 1467015 94 - 3904529 GGAAACAUUUCUGGUCAGAGAUAAUAAUUUUGCUUUAUUA-UUUCUGUUUU--UUCUCUCUUUUUUUGCUCU-------------CAUUCUGUCAUAGUGG--UGUGUUCAU--- (((.((((..(((..((((((((((((.......))))))-))))))....--..............((...-------------......))..)))..)--))).)))..--- ( -16.40, z-score = -1.84, R) >consensus UGAAACAUUUCUGGCC__AGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUU__CUGUCGUUCGGCCA_UUCUGUCC_________CAUCGAGUGCGAGUGGCCUUUGUUGAUGGU ...((((.....((((..(((((((((.......)))))))))...))))............(((((.(((........................))))))))..))))...... (-13.25 = -13.55 + 0.29)
| Location | 15,446,475 – 15,446,572 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Shannon entropy | 0.37936 |
| G+C content | 0.39893 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -13.32 |
| Energy contribution | -14.36 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
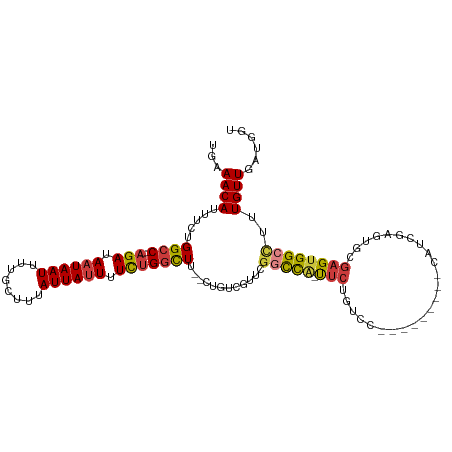
>dm3.chr2R 15446475 97 + 21146708 --------GAAUGGCCGAA--CGACAGAAGCCAAAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGGG-CCCCCAGAAAA --------....((((((.--.((((...((((.(.((((((........)))))).).))))......)))))).....((((.....)))).))-))......... ( -19.10, z-score = -1.48, R) >droSim1.chr2R 14147807 97 + 19596830 --------GAAUGGCCGAA--CGACAGAAGCCAGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGGG-CCCCCAGAAAA --------....((((((.--.((((...((((((.((((((........)))))).))))))......)))))).....((((.....)))).))-))......... ( -25.30, z-score = -3.67, R) >droSec1.super_1 12965915 97 + 14215200 --------GCAUGGCCGAA--CGACAGAAGCCAGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGGG-CCCCCAGAAAA --------....((((((.--.((((...((((((.((((((........)))))).))))))......)))))).....((((.....)))).))-))......... ( -25.30, z-score = -3.34, R) >droYak2.chr2R 7411404 98 - 21139217 --------GAAUGGCCGUA--CGACACAAGCCCGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGGGGCCCCCAGAAAA --------....((((...--((((....(((.((.((((((........)))))).)).)))......((((.....)))).......))))..))))......... ( -21.00, z-score = -1.15, R) >droEre2.scaffold_4845 9645843 96 + 22589142 --------GAAUGGCCGAA--CGACAGAAGCCAGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGG--CCCCCAGAAAA --------....((((.((--(.......((((((.((((((........)))))).))))))......((((.....)))).......))).))--))......... ( -26.00, z-score = -4.56, R) >droAna3.scaffold_13266 10053303 91 - 19884421 GAUGAUGGAGGCAGACAAGCGACUCUGAAGCCAGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAGGCGAAAGA----------------- .......((((((..........((((..((((((.((((((........)))))).))))))))))..)))))).........(....).----------------- ( -20.30, z-score = -2.21, R) >droPer1.super_2 30744 80 - 9036312 --------GGAGAGACGACUCCGACUAAGGCCACAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUACCAG-AGACA------------------- --------((((......))))......(((((.(.((((((........)))))).).))))).....((((((.......)-)))))------------------- ( -20.00, z-score = -4.24, R) >consensus ________GAAUGGCCGAA__CGACAGAAGCCAGAAAAUAAUAAAGCAAAAUUAUUAUCUGGCCAGAAAUGUUUCAAUAACAACAGACAGUUGGGG_CCCCCAGAAAA .............................((((((.((((((........)))))).))))))......((((.....))))........(((((....))))).... (-13.32 = -14.36 + 1.04)


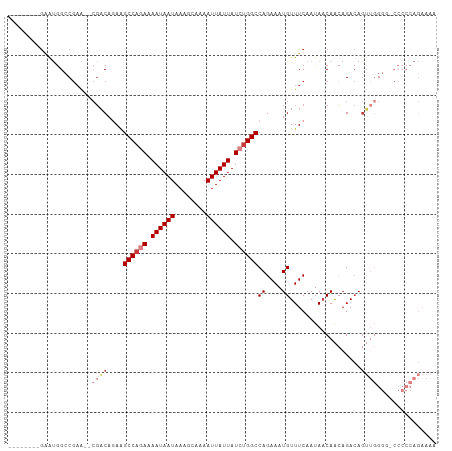
| Location | 15,446,475 – 15,446,572 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Shannon entropy | 0.37936 |
| G+C content | 0.39893 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
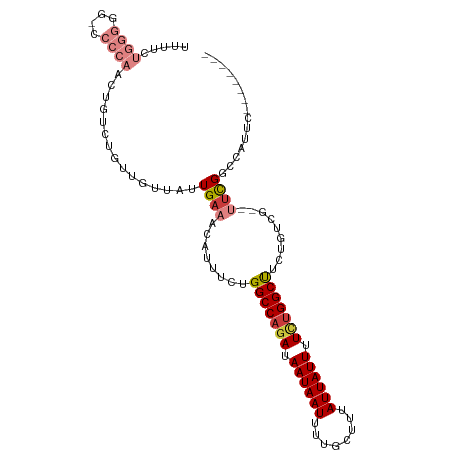
>dm3.chr2R 15446475 97 - 21146708 UUUUCUGGGGG-CCCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUUUGGCUUCUGUCG--UUCGGCCAUUC-------- .........((-((.((((.....)))).....(((........(((((((.((((((........)))))).))))))).......--)))))))....-------- ( -24.46, z-score = -2.12, R) >droSim1.chr2R 14147807 97 - 19596830 UUUUCUGGGGG-CCCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCUGUCG--UUCGGCCAUUC-------- .........((-((.((((.....)))).....(((........(((((((.((((((........)))))).))))))).......--)))))))....-------- ( -27.06, z-score = -3.10, R) >droSec1.super_1 12965915 97 - 14215200 UUUUCUGGGGG-CCCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCUGUCG--UUCGGCCAUGC-------- .........((-((.((((.....)))).....(((........(((((((.((((((........)))))).))))))).......--)))))))....-------- ( -27.06, z-score = -2.64, R) >droYak2.chr2R 7411404 98 + 21139217 UUUUCUGGGGGCCCCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCGGGCUUGUGUCG--UACGGCCAUUC-------- .........((((..((((.....))))....((..((((....((((.((.((((((........)))))).)).)))).))))..--)).))))....-------- ( -23.00, z-score = -0.64, R) >droEre2.scaffold_4845 9645843 96 - 22589142 UUUUCUGGGGG--CCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCUGUCG--UUCGGCCAUUC-------- .........((--((((((.....)))).....(((........(((((((.((((((........)))))).))))))).......--)))))))....-------- ( -27.06, z-score = -3.30, R) >droAna3.scaffold_13266 10053303 91 + 19884421 -----------------UCUUUCGCCUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCAGAGUCGCUUGUCUGCCUCCAUCAUC -----------------...............(((.....(((((((((((.((((((........)))))).))))))..)))))..((......)).....))).. ( -18.90, z-score = -2.08, R) >droPer1.super_2 30744 80 + 9036312 -------------------UGUCU-CUGGUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUGUGGCCUUAGUCGGAGUCGUCUCUCC-------- -------------------.(.((-((((....(((....))).(((((.(.((((((........)))))).).)))))....)))))).)........-------- ( -19.50, z-score = -2.37, R) >consensus UUUUCUGGGGG_CCCCAACUGUCUGUUGUUAUUGAAACAUUUCUGGCCAGAUAAUAAUUUUGCUUUAUUAUUUUCUGGCUUCUGUCG__UUCGGCCAUUC________ .....((((....))))...........................(((((((.((((((........)))))).)))))))...((((....))))............. (-15.70 = -16.27 + 0.57)

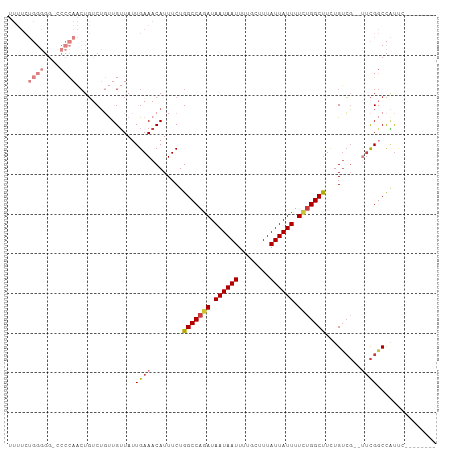
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:07 2011