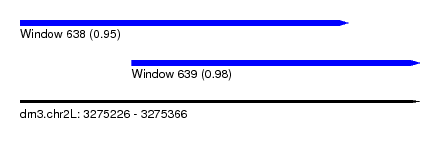
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,275,226 – 3,275,366 |
| Length | 140 |
| Max. P | 0.983896 |

| Location | 3,275,226 – 3,275,341 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 62.54 |
| Shannon entropy | 0.74828 |
| G+C content | 0.48263 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -8.59 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
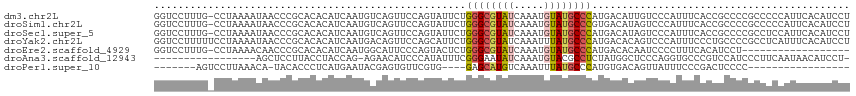
>dm3.chr2L 3275226 115 + 23011544 GGUCCUUUG-CCUAAAAUAACCCGCACACAUCAAUGUCAGUUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACAUUGUCCCAUUUCACCGCCCCGCCCCCAUUCACAUCCU (((....((-(............))).....((((((((.....((....))((((((((.....)))))))).)))))))).........)))...................... ( -22.10, z-score = -1.86, R) >droSim1.chr2L 3212096 115 + 22036055 GGUCCUUUG-CCUAAAAUAACCCGCACACAUCAAUGUCAGUUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCGUGACAUAGUCCCAUUUCACCGCCCCGCCCCCAUUCACAUCCU ((..((.((-(............))).......((((((.....((....))((((((((.....)))))))).))))))))..)).............................. ( -19.60, z-score = -0.88, R) >droSec1.super_5 1417612 115 + 5866729 GGUCCUUUG-CCUAAAAUAACCCGCACACAUCAAUGUCAGUUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACAUAGUCCCAUUUCACCGCCCCGCCUCCAUUCACAUCCU ((..((.((-(............))).......((((((.....((....))((((((((.....)))))))).))))))))..)).............................. ( -19.70, z-score = -1.13, R) >droYak2.chr2L 3258533 116 + 22324452 GGUCCUUUUUCCUAAAAUAACCCGCACACAUCAAUGACAGUUCCAGCAUUCUGGGCGUAUCAAAUUUAUGCCCAUGACACAGUCCCAUUUCCCUGCCCCGCCUCAUUUCACAUCCU (((..((((....))))..))).(((.........(((.((....))..((((((((((.......)))))))).))....))).........))).................... ( -18.57, z-score = -1.41, R) >droEre2.scaffold_4929 3306603 97 + 26641161 GGUCCUUUG-CCUAAAACAACCCGCACACAUCAAUGGCAUUCCCAGUACUCUGGGCGUAUCAAAUGUAUGCCCAUGACACAAUCCCCUUUCACAUCCU------------------ .(((...((-((.......................))))............(((((((((.....))))))))).)))....................------------------ ( -18.40, z-score = -1.07, R) >droAna3.scaffold_12943 30875 97 + 5039921 -----------------AGCUCCUUACCUACCAG-AGAACAUCCCAUAUUUCGGGAAUAUCAAAUGUACGCCUCUAUGGCUCCCAGGUGCCCGUCCAUCCCUUCAAUAACAUCCU- -----------------.((.(((..........-.((...((((.......))))...))........(((.....)))....))).)).........................- ( -15.50, z-score = -0.04, R) >droPer1.super_10 3091120 87 - 3432795 -------AGUCCUUAAACA-UACACCCUCAUGAAUACGAGUGUUCGUG----GAGCAUGUCAAAUUUAUGCCCAUGUGACAGUUAUUUCCCGACUCCCC----------------- -------((((........-......(((........)))((((((((----(.(((((.......)))))))))).))))..........))))....----------------- ( -18.40, z-score = -1.89, R) >consensus GGUCCUUUG_CCUAAAAUAACCCGCACACAUCAAUGUCAGUUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACACAGUCCCAUUUCACCGCCCCGCCUCCAUUCACAUCCU ....................................................((((((((.....))))))))........................................... ( -8.59 = -8.96 + 0.37)

| Location | 3,275,265 – 3,275,366 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 57.78 |
| Shannon entropy | 0.80157 |
| G+C content | 0.48160 |
| Mean single sequence MFE | -14.99 |
| Consensus MFE | -7.86 |
| Energy contribution | -7.91 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3275265 101 + 23011544 UUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACAUUGUCCCAUUUCACCGCCCCGCCCCCAUUCACAUCCUUUCAAUAGUCUACCUUUUUU-GACAU------------- ...((((.((.(((((((((.....))))))))).)).)))).........................................(((..........-)))..------------- ( -16.90, z-score = -2.09, R) >droSim1.chr2L 3212135 101 + 22036055 UUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCGUGACAUAGUCCCAUUUCACCGCCCCGCCCCCAUUCACAUCCUUUCAAUAGCCUCCCUUUUUU-GACAU------------- ............((((((((.....))))))))((((.((......)).))))........................((((.((.....))...))-))...------------- ( -13.80, z-score = -0.75, R) >droSec1.super_5 1417651 101 + 5866729 UUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACAUAGUCCCAUUUCACCGCCCCGCCUCCAUUCACAUCCUAUCAAUAGCCUCCCUUUUUU-GACAU------------- .........(((((((((((.....))))))))).))....(((..........((...)).............(((....)))............-)))..------------- ( -13.20, z-score = -0.53, R) >droYak2.chr2L 3258573 102 + 22324452 UUCCAGCAUUCUGGGCGUAUCAAAUUUAUGCCCAUGACACAGUCCCAUUUCCCUGCCCCGCCUCAUUUCACAUCCUUUCAAUAGCCGCCCUUUUUUUGACAU------------- .....((..((((((((((.......)))))))).))..(((..........)))....)).........................................------------- ( -13.70, z-score = -0.57, R) >droEre2.scaffold_4929 3306642 82 + 26641161 UCCCAGUACUCUGGGCGUAUCAAAUGUAUGCCCAUGACACAAUCCCCUUUC------------------ACAUCCUUUCAAUAGCCGCCCUUUUU--GACAU------------- .....((..(((((((((((.....))))))))).)).))...........------------------........((((.((.....))..))--))...------------- ( -13.20, z-score = -1.25, R) >droAna3.scaffold_12943 30899 112 + 5039921 --CCCAUAUUUCGGGAAUAUCAAAUGUACGCCUCU-AUGGCUCCCAGGUGCCCGUCCAUCCCUUCAAUAACAUCCUUUCGAUGUCAAUCCGCUCUCCGCCUCCGCAAAUUGUCAU --..........((((.............(((...-..))).....((.......)).))))..((((.(((((.....))))).............((....))..)))).... ( -16.30, z-score = 0.44, R) >dp4.chr4_group4 1330539 78 + 6586962 ----UGUUCGUGGAGCAUGUCAAAUUUAUGCCCACGUGACAGUU-UUUCCCGACUCCCCC-UGCCAGCUCCACCAUCUGUACAU------------------------------- ----.....(((((((.(((((..............)))))(((-......)))......-.....)))))))...........------------------------------- ( -16.34, z-score = -1.61, R) >droPer1.super_10 3091152 80 - 3432795 ----UGUUCGUGGAGCAUGUCAAAUUUAUGCCCAUGUGACAGUUAUUUCCCGACUCCCCCCUGCCAGCUCCACCAUCUGUACAU------------------------------- ----.....(((((((.(((((.((........)).)))))(((.......)))............)))))))...........------------------------------- ( -16.50, z-score = -1.61, R) >consensus UUCCAGUAUUCUGGGCGUAUCAAAUGUAUGCCCAUGACACAGUCCCAUUUCACCGCCCCGCCUCCAUUCACAUCCUUUCAAUAGCCUCCCUUUUUU_GACAU_____________ ............(((((((.......))))))).................................................................................. ( -7.86 = -7.91 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:32 2011