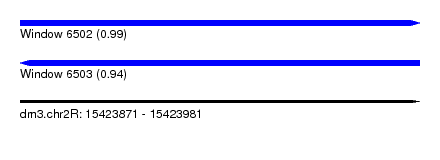
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,423,871 – 15,423,981 |
| Length | 110 |
| Max. P | 0.991209 |

| Location | 15,423,871 – 15,423,981 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.88 |
| Shannon entropy | 0.48025 |
| G+C content | 0.33328 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
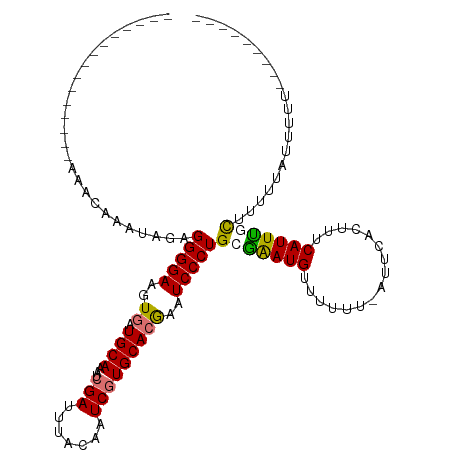
>dm3.chr2R 15423871 110 + 21146708 AAAAAAAAAAGUGAAAAACAUACAGAGGGGAAGUGAUGCAAAUCGAUUUACAAUCAUGCACGAAUCCCUGCGAAUGUUUUUU-AUUUACUUUCAUUUGCUUUUUAUAUUUA-------- ........(((((((((((((.(...(((((..((.((((....(((.....))).))))))..)))))..).)))))))))-))))........................-------- ( -22.90, z-score = -2.02, R) >droEre2.scaffold_4845 9623384 97 + 22589142 -------------------AAAU-GAGGGGAAGUGAUGCAAAUCGAUUUACAUUCGUGCACAAAUCCCUG-GGAUGUUUUUU-AUUCACUUCCAUUCGCUUUGUAUUUUCUGCUACUCA -------------------...(-(((.((((((((((((...(((.......)))))))...((((...-)))).......-..))))))))....((............))..)))) ( -22.40, z-score = -1.44, R) >droYak2.chr2R 7388178 87 - 21139217 ---------------GAAAAAAUAAAGGGGAAGUCAUGCAAAUCGAUUUACAUUCGUGCAUGAAUCCCUGAGGAUGUUUUUUUAUUCGCUUUCAUUUGUAAA----------------- ---------------((((.......(((((..(((((((...(((.......)))))))))).)))))..(((((......)))))..)))).........----------------- ( -22.20, z-score = -2.44, R) >droSim1.chr2R 14124962 113 + 19596830 ----AAAAAAGAGAAAAACAAAUAGAGGGGAAGUGAUGCAAAUCGAUUUACAAUCGUGCACGAAUCCCUGCAAAUGUUUUUU-CUUCACUUUCAUUUGCUUUUUAUUUUUGGUU-CUCA ----......(((((..(.((((((((((((..((.((((...((((.....))))))))))..)))))(((((((......-.........)))))))..))))))).)..))-))). ( -30.16, z-score = -3.00, R) >consensus _______________AAACAAAUAGAGGGGAAGUGAUGCAAAUCGAUUUACAAUCGUGCACGAAUCCCUGCGAAUGUUUUUU_AUUCACUUUCAUUUGCUUUUUAUUUUU_________ ..........................(((((..((.((((...(((.......)))))))))..)))))(((((((................))))))).................... (-15.24 = -15.24 + 0.00)

| Location | 15,423,871 – 15,423,981 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.88 |
| Shannon entropy | 0.48025 |
| G+C content | 0.33328 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -12.00 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
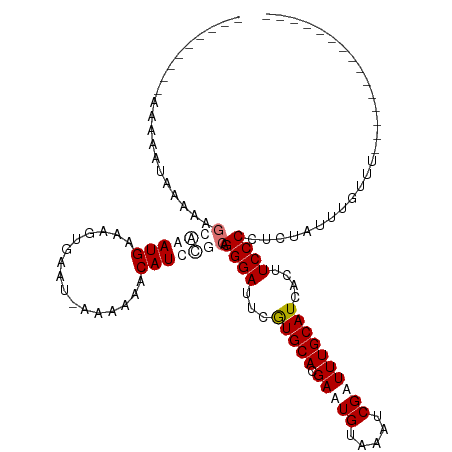
>dm3.chr2R 15423871 110 - 21146708 --------UAAAUAUAAAAAGCAAAUGAAAGUAAAU-AAAAAACAUUCGCAGGGAUUCGUGCAUGAUUGUAAAUCGAUUUGCAUCACUUCCCCUCUGUAUGUUUUUCACUUUUUUUUUU --------..............(((.((((((....-.((((((((.((.((((....(((((.(((((.....))))))))))......)))).)).)))))))).)))))).))).. ( -20.90, z-score = -1.85, R) >droEre2.scaffold_4845 9623384 97 - 22589142 UGAGUAGCAGAAAAUACAAAGCGAAUGGAAGUGAAU-AAAAAACAUCC-CAGGGAUUUGUGCACGAAUGUAAAUCGAUUUGCAUCACUUCCCCUC-AUUU------------------- ((((..((............))....((((((((..-.......((((-...))))...(((((((.......)))...)))))))))))).)))-)...------------------- ( -19.70, z-score = -1.14, R) >droYak2.chr2R 7388178 87 + 21139217 -----------------UUUACAAAUGAAAGCGAAUAAAAAAACAUCCUCAGGGAUUCAUGCACGAAUGUAAAUCGAUUUGCAUGACUUCCCCUUUAUUUUUUC--------------- -----------------..................................((((.((((((((((.......)))...)))))))..))))............--------------- ( -15.90, z-score = -1.68, R) >droSim1.chr2R 14124962 113 - 19596830 UGAG-AACCAAAAAUAAAAAGCAAAUGAAAGUGAAG-AAAAAACAUUUGCAGGGAUUCGUGCACGAUUGUAAAUCGAUUUGCAUCACUUCCCCUCUAUUUGUUUUUCUCUUUUUU---- .(((-((....(((((((..(((((((.........-......))))))).((((...(((((.(((((.....))))))))))....)))).....))))))))))))......---- ( -26.36, z-score = -2.37, R) >consensus _________AAAAAUAAAAAGCAAAUGAAAGUGAAU_AAAAAACAUCCGCAGGGAUUCGUGCACGAAUGUAAAUCGAUUUGCAUCACUUCCCCUCUAUUUGUUU_______________ ....................(((((((................))))))).((((...(((((.(((((.....))))))))))....))))........................... (-12.00 = -13.94 + 1.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:35:01 2011