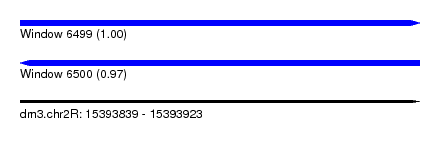
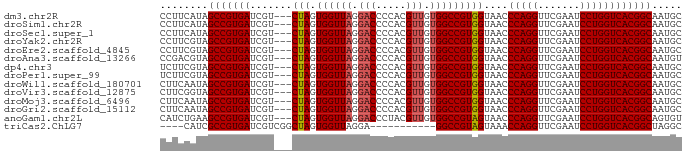
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,393,839 – 15,393,923 |
| Length | 84 |
| Max. P | 0.995261 |

| Location | 15,393,839 – 15,393,923 |
|---|---|
| Length | 84 |
| Sequences | 14 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Shannon entropy | 0.18596 |
| G+C content | 0.57143 |
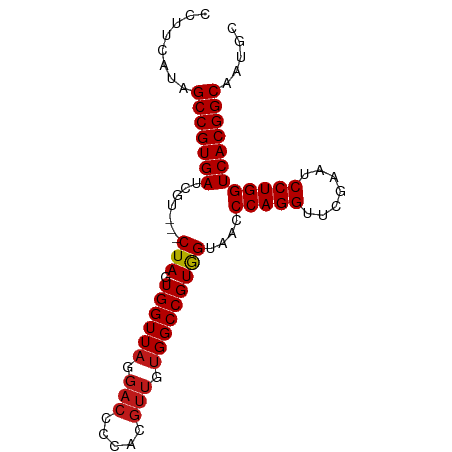
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -28.08 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15393839 84 + 21146708 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUGAAGG .(((.((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..))))))).))).... ( -33.90, z-score = -3.31, R) >droSim1.chr2R 14092707 84 + 19596830 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUGAAGG .(((.((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..))))))).))).... ( -33.90, z-score = -3.31, R) >droSec1.super_1 12912648 84 + 14215200 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUGAAGG .(((.((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..))))))).))).... ( -33.90, z-score = -3.31, R) >droYak2.chr2R 7357588 84 - 21139217 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACGAAGG .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.62, R) >droEre2.scaffold_4845 9593201 84 + 22589142 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACGAAGG .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.62, R) >droAna3.scaffold_13266 14368661 84 - 19884421 ACAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACGUCGG .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.19, R) >dp4.chr3 12520878 84 + 19779522 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACGAAGA .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.78, R) >droPer1.super_99 151300 84 - 176816 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACGAAGA .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.78, R) >droWil1.scaffold_180701 1374974 84 + 3904529 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUUGAAG .((..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))...))... ( -32.30, z-score = -3.22, R) >droVir3.scaffold_12875 16266562 84 - 20611582 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUACCGAAG .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -31.70, z-score = -2.82, R) >droMoj3.scaffold_6496 20696146 84 + 26866924 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUUGAAG .((..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))...))... ( -32.30, z-score = -3.22, R) >droGri2.scaffold_15112 1906239 84 + 5172618 GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG---ACGAUCACGGCUAUUGAAG .((..((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))...))... ( -32.30, z-score = -3.22, R) >anoGam1.chr2L 19552914 84 - 48795086 ACACUGCCGUGACCAGGAUUCGAACCUGGGUUACUACGGCCACAACGUAGGGUCCUAACCACUAG---ACGAUCACGGCUUCAGAUG .....((((((((((((.......)))))(((.(((((.......)))))(((....)))....)---))..)))))))........ ( -29.70, z-score = -2.96, R) >triCas2.ChLG7 1603135 72 + 17478683 GCCUAGCCGUGACCAGGAUUCGAACCUGGUUUACUACGGCC-----------UCCUAACCACUAGCCGACGAUCACGGCGAUG---- .....((((((((((((.......))))).......((((.-----------............))))....)))))))....---- ( -25.42, z-score = -3.66, R) >consensus GCAUUGCCGUGACCAGGAUUCGAACCUGGGUUACCACGGCCACAACGUGGGGUCCUAACCACUAG___ACGAUCACGGCUACGAAGG .....((((((((((((.......)))))....(((((.......)))))(((....)))............)))))))........ (-28.08 = -28.46 + 0.38)
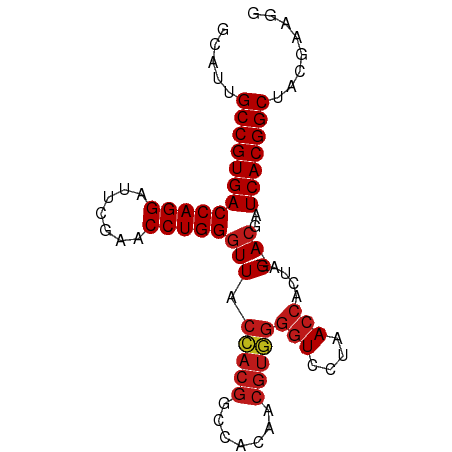
| Location | 15,393,839 – 15,393,923 |
|---|---|
| Length | 84 |
| Sequences | 14 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Shannon entropy | 0.18596 |
| G+C content | 0.57143 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15393839 84 - 21146708 CCUUCAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.70, z-score = -1.84, R) >droSim1.chr2R 14092707 84 - 19596830 CCUUCAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.70, z-score = -1.84, R) >droSec1.super_1 12912648 84 - 14215200 CCUUCAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.70, z-score = -1.84, R) >droYak2.chr2R 7357588 84 + 21139217 CCUUCGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.30, z-score = -1.53, R) >droEre2.scaffold_4845 9593201 84 - 22589142 CCUUCGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.30, z-score = -1.53, R) >droAna3.scaffold_13266 14368661 84 + 19884421 CCGACGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGU ...((((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).)))) ( -31.90, z-score = -1.64, R) >dp4.chr3 12520878 84 - 19779522 UCUUCGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.30, z-score = -1.63, R) >droPer1.super_99 151300 84 + 176816 UCUUCGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ....(((.(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......)))))))))))).))). ( -30.30, z-score = -1.63, R) >droWil1.scaffold_180701 1374974 84 - 3904529 CUUCAAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ........(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... ( -30.00, z-score = -1.82, R) >droVir3.scaffold_12875 16266562 84 + 20611582 CUUCGGUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ........(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... ( -30.00, z-score = -1.14, R) >droMoj3.scaffold_6496 20696146 84 - 26866924 CUUCAAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ........(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... ( -30.00, z-score = -1.82, R) >droGri2.scaffold_15112 1906239 84 - 5172618 CUUCAAUAGCCGUGAUCGU---CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ........(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... ( -30.00, z-score = -1.82, R) >anoGam1.chr2L 19552914 84 + 48795086 CAUCUGAAGCCGUGAUCGU---CUAGUGGUUAGGACCCUACGUUGUGGCCGUAGUAACCCAGGUUCGAAUCCUGGUCACGGCAGUGU ........(((((((....---(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... ( -30.30, z-score = -2.14, R) >triCas2.ChLG7 1603135 72 - 17478683 ----CAUCGCCGUGAUCGUCGGCUAGUGGUUAGGA-----------GGCCGUAGUAAACCAGGUUCGAAUCCUGGUCACGGCUAGGC ----....(((((((......((((.(((((....-----------)))))))))...(((((.......))))))))))))..... ( -27.20, z-score = -2.11, R) >consensus CCUUCAUAGCCGUGAUCGU___CUAGUGGUUAGGACCCCACGUUGUGGCCGUGGUAACCCAGGUUCGAAUCCUGGUCACGGCAAUGC ........(((((((.......(((.((((((.(((.....))).)))))))))....(((((.......))))))))))))..... (-29.30 = -29.41 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:58 2011