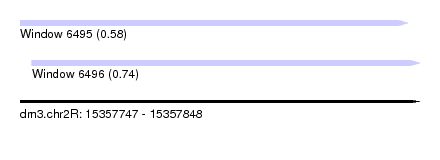
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,357,747 – 15,357,848 |
| Length | 101 |
| Max. P | 0.742082 |

| Location | 15,357,747 – 15,357,845 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.72 |
| Shannon entropy | 0.66618 |
| G+C content | 0.57091 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.26 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
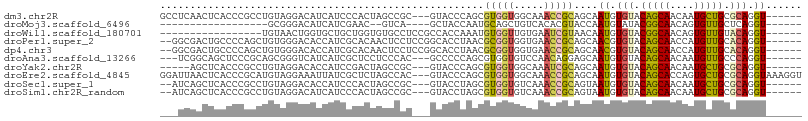
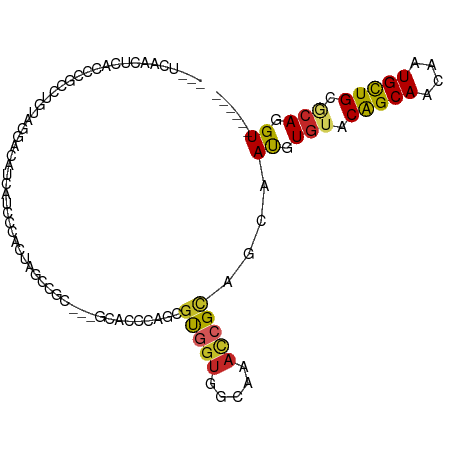
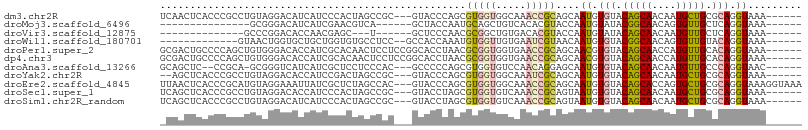
>dm3.chr2R 15357747 98 + 21146708 GCCUCAACUCACCCGCCUGUAGGACAUCAUCCCACUAGCCGC---GUACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCAACAAUGCUGCGCAGGU------ ((((..........(((.((.(((.....))).))..(((((---((.....))))))))))....(((((((.(((........))).))))))).))))------ ( -35.10, z-score = -1.77, R) >droMoj3.scaffold_6496 15062993 77 + 26866924 ------------------GCGGGACAUCAUCGAAC--GUCA----GCUACCAAUGCAGCUGUCACACGUACCAAUGUAUACGGCAACAGUGUUGCUCAGGU------ ------------------((..(((..........--))).----)).(((...((((((((..(.((((........)))))..)))))..)))...)))------ ( -16.80, z-score = 0.94, R) >droWil1.scaffold_180701 1325762 84 + 3904529 -----------------UGUAACUGGUGCUGCUGGUGUGCCUCCGCCACCAAAUGUGGUUGUGAAUCGUAACAAUGUGUACGGCAACAGUGUUGUACAGGU------ -----------------(((.((.((..(.......)..))..(((.((((....)))).)))....)).)))((.(((((((((....))))))))).))------ ( -24.60, z-score = 0.25, R) >droPer1.super_2 2320285 99 - 9036312 --GGCGACUGCCCCAGCUGUGGGACACCAUCGCACAACUCCUCCGGCACCUAACGCGGUGGUGAACCGCAGCAACGUGUACAGCAACCAUGUUGCACAGGU------ --(((....)))...(((((((..(((((((((.....................)))))))))..))))))).((.(((.(((((....))))).))).))------ ( -41.00, z-score = -1.99, R) >dp4.chr3 13283111 99 - 19779522 --GGCGACUGCCCCAGCUGUGGGACACCAUCGCACAACUCCUCCGGCACCUAACGCGGUGGUGAACCGCAGCAACGUGUACAGCAACCAUGUUGCACAGGU------ --(((....)))...(((((((..(((((((((.....................)))))))))..))))))).((.(((.(((((....))))).))).))------ ( -41.00, z-score = -1.99, R) >droAna3.scaffold_13266 9962013 95 - 19884421 ---UCGGCAGCUCCCGCAGCGGGUCAUCAUCGCUCCUCCCAC---GCCCCCAGCGUGGUGUCCAACAGGAGCAAUGUGUACAGCAACAAUGUUGCCCAGGU------ ---..((((((.((((...)))).((.(((.(((((((((((---((.....)))))).)......)))))).)))))............)))))).....------ ( -33.60, z-score = -1.59, R) >droYak2.chr2R 7320772 93 - 21139217 -----AGCUCACCCGCCUGUAGGACACCAUCCGACUAGCCGC---GUACCCAGCGUGGUGGCAAAUCGCAGCAAUGUGUACAGCAACAAUGCUGCGCAGGU------ -----.....(((.(((.((.(((.....))).))..(((((---((.....))))))))))....(((((((.(((........))).)))))))..)))------ ( -33.90, z-score = -1.17, R) >droEre2.scaffold_4845 9556482 104 + 22589142 GGAUUAACUCACCCGCAUGUAGGAAAUUAUCGCUCUAGCCAC---GUACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCACCAGUGCUGCGCAGGUAAAGGU ..........(((.((...............(((...(((((---((.....)))))))))).....((((((..(((.....)))...)))))))).)))...... ( -33.70, z-score = -0.64, R) >droSec1.super_1 12876230 96 + 14215200 --AUCAGCUCACCCGCCUGUAGGACACCAUCCCACUAGCCGC---GUACCUAGCGUGGUGUCAAACCGCAGUAAUGUGUACAGCAACAAUGCUGCGCAGGU------ --............((((((..((((((((....((((....---....)))).)))))))).....((((((.(((........))).))))))))))))------ ( -33.70, z-score = -2.05, R) >droSim1.chr2R_random 2645700 96 + 2996586 --AUCAGCUCACCCGCCUGUAGGACAUCAUCCCACUAGCCGC---GUACCUAGCGUGGUGUCAAACCGCAGUAAUGUGUACAGCAACAAUGCUGCGCAGGU------ --............((((((..((((((((....((((....---....)))).)))))))).....((((((.(((........))).))))))))))))------ ( -31.10, z-score = -1.40, R) >consensus ___UCAACUCACCCGCCUGUAGGACAUCAUCCCACUAGCCGC___GCACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCAACAAUGCUGCGCAGGU______ ......................................................(((((.....)))))....((.(((.(((((....))))).))).))...... (-12.67 = -12.26 + -0.41)
| Location | 15,357,750 – 15,357,848 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.29 |
| Shannon entropy | 0.68110 |
| G+C content | 0.55188 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -12.09 |
| Energy contribution | -11.99 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15357750 98 + 21146708 UCAACUCACCCGCCUGUAGGACAUCAUCCCACUAGCCGC---GUACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCAACAAUGCUGCGCAGGUAAA------ .......(((.(((.((.(((.....))).))..(((((---((.....))))))))))....(((((((.(((........))).)))))))..)))...------ ( -34.80, z-score = -2.21, R) >droMoj3.scaffold_6496 15062993 80 + 26866924 ---------------GCGGGACAUCAUCGAACGUCA------GCUACCAAUGCAGCUGUCACACGUACCAAUGUAUACGGCAACAGUGUUGCUCAGGUAAA------ ---------------((..(((..........))).------))((((...((((((((..(.((((........)))))..)))))..)))...))))..------ ( -18.80, z-score = 0.20, R) >droVir3.scaffold_12875 8813062 78 - 20611582 --------------GCCCGGACACCAACGAGC---U------GCUCCCAACGCGGCUGUGACACGUACCAAUGUAUACAGCAACAAUGUUGCUCAGGUAAA------ --------------(((.((((....((.(((---(------((.......)))))))).....)).)).........((((((...))))))..)))...------ ( -19.30, z-score = -0.02, R) >droWil1.scaffold_180701 1325763 86 + 3904529 -------------GUAACUGGUGCUGCUGGUGUGCCUCC--GCCACCAAAUGUGGUUGUGAAUCGUAACAAUGUGUACGGCAACAGUGUUGUACAGGUAAA------ -------------((.((.((..(.......)..))..(--((.((((....)))).)))....)).)).((.(((((((((....))))))))).))...------ ( -24.60, z-score = 0.13, R) >droPer1.super_2 2320286 101 - 9036312 GCGACUGCCCCAGCUGUGGGACACCAUCGCACAACUCCUCCGGCACCUAACGCGGUGGUGAACCGCAGCAACGUGUACAGCAACCAUGUUGCACAGGUAAA------ .....((((...(((((((..(((((((((.....................)))))))))..)))))))....(((.(((((....))))).)))))))..------ ( -39.10, z-score = -1.95, R) >dp4.chr3 13283112 101 - 19779522 GCGACUGCCCCAGCUGUGGGACACCAUCGCACAACUCCUCCGGCACCUAACGCGGUGGUGAACCGCAGCAACGUGUACAGCAACCAUGUUGCACAGGUAAA------ .....((((...(((((((..(((((((((.....................)))))))))..)))))))....(((.(((((....))))).)))))))..------ ( -39.10, z-score = -1.95, R) >droAna3.scaffold_13266 9962016 95 - 19884421 GCAGCUC--CCGCA-GCGGGUCAUCAUCGCUCCUCCCAC---GCCCCCAGCGUGGUGUCCAACAGGAGCAAUGUGUACAGCAACAAUGUUGCCCAGGUAAC------ ((.((..--..)).-))((((((.(((.(((((((((((---((.....)))))).)......)))))).))))).)).(((((...))))))).......------ ( -31.20, z-score = -1.48, R) >droYak2.chr2R 7320772 96 - 21139217 --AGCUCACCCGCCUGUAGGACACCAUCCGACUAGCCGC---GUACCCAGCGUGGUGGCAAAUCGCAGCAAUGUGUACAGCAACAAUGCUGCGCAGGUAAA------ --.....(((.(((.((.(((.....))).))..(((((---((.....))))))))))....(((((((.(((........))).)))))))..)))...------ ( -34.60, z-score = -1.44, R) >droEre2.scaffold_4845 9556485 104 + 22589142 UUAACUCACCCGCAUGUAGGAAAUUAUCGCUCUAGCCAC---GUACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCACCAGUGCUGCGCAGGUAAAGGUAAA ...(((.(((.((...............(((...(((((---((.....)))))))))).....((((((..(((.....)))...)))))))).)))...)))... ( -33.90, z-score = -1.10, R) >droSec1.super_1 12876231 98 + 14215200 UCAGCUCACCCGCCUGUAGGACACCAUCCCACUAGCCGC---GUACCUAGCGUGGUGUCAAACCGCAGUAAUGUGUACAGCAACAAUGCUGCGCAGGUAAA------ ...........((((((..((((((((....((((....---....)))).)))))))).....((((((.(((........))).))))))))))))...------ ( -34.40, z-score = -2.36, R) >droSim1.chr2R_random 2645701 98 + 2996586 UCAGCUCACCCGCCUGUAGGACAUCAUCCCACUAGCCGC---GUACCUAGCGUGGUGUCAAACCGCAGUAAUGUGUACAGCAACAAUGCUGCGCAGGUAAA------ ...........((((((..((((((((....((((....---....)))).)))))))).....((((((.(((........))).))))))))))))...------ ( -31.80, z-score = -1.71, R) >consensus _CAACUCACCCGCCUGUAGGACACCAUCGCACUACCCGC___GCACCCAGCGUGGUGGCAAACCGCAGCAAUGUGUACAGCAACAAUGUUGCGCAGGUAAA______ ...................................................(((((.....)))))....((.(((.(((((....))))).))).))......... (-12.09 = -11.99 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:55 2011