| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,354,733 – 15,354,800 |
| Length | 67 |
| Max. P | 0.955889 |

| Location | 15,354,733 – 15,354,800 |
|---|---|
| Length | 67 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Shannon entropy | 0.60555 |
| G+C content | 0.46662 |
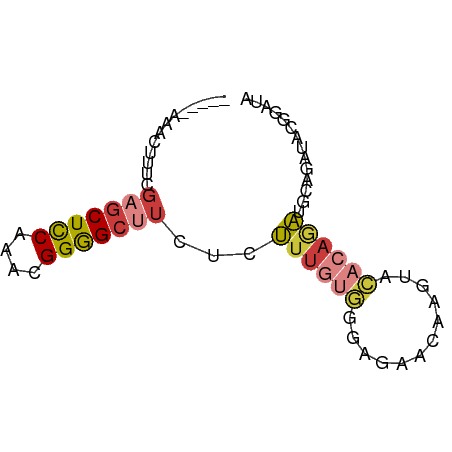
| Mean single sequence MFE | -15.94 |
| Consensus MFE | -8.23 |
| Energy contribution | -9.01 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
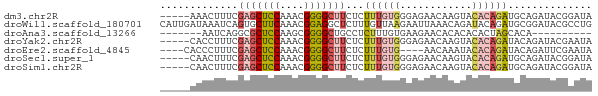
>dm3.chr2R 15354733 67 + 21146708 -----AAACUUUCGAGCUCCAAACGGGGCUUCUCUUUGUGGGAGAACAAGUACACAGAUGCAGAUACGGAUA -----.....((((((((((....))))))((((((((((............)))))).).)))..)))).. ( -16.50, z-score = -1.22, R) >droWil1.scaffold_180701 1322061 72 + 3904529 CAUUGAUAAAUCAGUGCUUCAAACGGAGGCUCUUUGUUAAGAAUUAAACAGAUACAGAUGCGGAUACGCCUG ..(((((((...((.(((((.....))))).)))))))))..............(((..(((....)))))) ( -14.10, z-score = -0.92, R) >droAna3.scaffold_13266 9958843 55 - 19884421 -------AAUCAGGCGCUCCAAGCGGGGCUGCCUCUUUGUGAAGAACACACACACUAGCACA---------- -------....(((((((((....))))).))))...((((.....))))............---------- ( -15.60, z-score = -1.09, R) >droYak2.chr2R 7317731 67 - 21139217 -----CACCUUUCGAGCUCCAAACGGGGCUUCUCUUUGUGGGAGAACAAGUACACAGAUACAGAUACGAAUA -----.....((((((((((....))))))(((.((((((............))))))...)))..)))).. ( -16.00, z-score = -1.50, R) >droEre2.scaffold_4845 9553403 64 + 22589142 ----CACCCUUUCGAGCUCCAAACGGGGCUUCUCUUUGUG----AACAAAUACACAGAUACAGAUUCGAAUA ----......((((((((((....))))).(((.((((((----........))))))...))).))))).. ( -16.40, z-score = -3.04, R) >droSec1.super_1 12873241 67 + 14215200 -----CAACUUUCGAGCUCCAAACGGGGCUUCUCUUUGUGGGAGAACAAGUACACAGAUGCAGAUACGGAUA -----.....((((((((((....))))))((((((((((............)))))).).)))..)))).. ( -16.50, z-score = -1.25, R) >droSim1.chr2R 14068713 67 + 19596830 -----CAACUUUCGAGCUCCAAACGGGGCUUCUCUUUGUGGGAGAACAAGUACACAGAUGCAGAUACGGAUA -----.....((((((((((....))))))((((((((((............)))))).).)))..)))).. ( -16.50, z-score = -1.25, R) >consensus _____AAACUUUCGAGCUCCAAACGGGGCUUCUCUUUGUGGGAGAACAAGUACACAGAUGCAGAUACGGAUA .............(((((((....)))))))...((((((............)))))).............. ( -8.23 = -9.01 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:53 2011