| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,329,738 – 15,329,876 |
| Length | 138 |
| Max. P | 0.989153 |

| Location | 15,329,738 – 15,329,876 |
|---|---|
| Length | 138 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 73.88 |
| Shannon entropy | 0.50717 |
| G+C content | 0.50432 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -23.82 |
| Energy contribution | -25.13 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
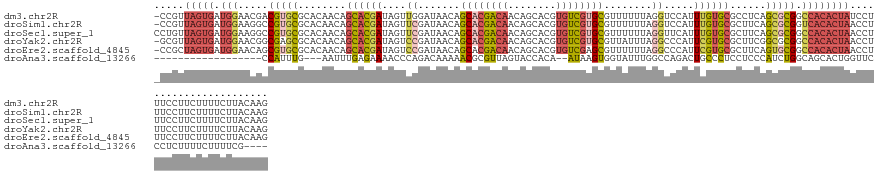
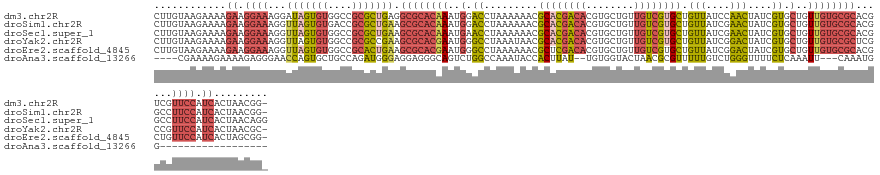
>dm3.chr2R 15329738 138 + 21146708 -CCGUUAGUGAUGGAACGACGUGCGCACAACAGCACGAUAGUUGGAUAACAGCACGACAACAGCACGUGUCGUGCGUUUUUUAGGUCCAUUUGUGCGCCUCAGCGCGGCCACACUAUCCUUUCCUUCUUUUCUUACAAG -..((.((.((.((((.(..((((........))))((((((((((((((.((((((((........)))))))))))......)))))...(((.(((.......))))))))))))).)))).))....)).))... ( -43.00, z-score = -1.99, R) >droSim1.chr2R 14042184 138 + 19596830 -CCGUUAGUGAUGGAAGGCCGUGCGCACAACAGCACGAUAGUUCGAUAACAGCACGACAACAGCACGUGUCGUGCGUUUUUUAGGUCCAUUUGUGCGCUUCAGCGCGGUCACACUAACCUUUCCUUCUUUUCUUACAAG -..(((((((.(((..(((.(.((((((((.....(((....)))......((((((((........))))))))...............))))))))..).)).)..))))))))))..................... ( -42.80, z-score = -2.11, R) >droSec1.super_1 12845753 139 + 14215200 CCUGUUAGUGAUGGAAGGCCGUGCGCACAACAGCACGAUAGUUCGAUAACAGCACGACAACAGCACGUGUCGUGCGUUUUUUAGGUUCAUUUGUGCGCUUCAGCGCGGCCACACUAACCUUUCCUUCUUUUCUUACAAG ...(((((((.(((..(((.(.((((((((.(((.(((....)))......((((((((........)))))))).........)))...))))))))..).)).)..))))))))))..................... ( -45.10, z-score = -2.55, R) >droYak2.chr2R 7292471 138 - 21139217 -GCGUUAGUGAUGGAACGGCGAGCGCACAACAGCACGAUAGUCCGAUAACAGCACGACAACAGCACGUGUCGUGCGUUAUUUAGGCCCAUUCGUGCGCUUCGGCGCGGCCACACUAACCUUUCCUUCUUUUCUUACAAG -..(((((((..(((....((.((........)).))....)))((((((.((((((((........))))))))))))))..((((.......((((....)))))))).)))))))..................... ( -49.61, z-score = -3.24, R) >droEre2.scaffold_4845 9528374 138 + 22589142 -CCGCUAGUGAUGGAACAGCGUGCGCACAACAGCACGAUAGUCCGAUAACAGCACGACAACAGCACGUGUCGAGCGUUUUUUAGGCCCAUUCGUGCGCUUCAGUGCGGCCACACUAACCUUUCCUUCUUUUCUUACAAG -....(((((.((((....(((((........)))))....))))......((.(((((........))))).))........((((.......((((....)))))))).)))))....................... ( -37.71, z-score = -0.79, R) >droAna3.scaffold_13266 17949152 112 - 19884421 ------------------CCAUUUG---AAUUUGAGAAAACCCAGACAAAAACGCGUUAGUACCACA--AUAAGUGGUAUUUGGCCAGACUGCCCUCCUCCCAUCUGGCAGCACUGGUUCCCUCUUUUCUUUUCG---- ------------------......(---((...(((..((((.((........((...((((((((.--....))))))))..))..(.(((((............)))))).))))))..)))..)))......---- ( -27.50, z-score = -2.15, R) >consensus _CCGUUAGUGAUGGAACGCCGUGCGCACAACAGCACGAUAGUCCGAUAACAGCACGACAACAGCACGUGUCGUGCGUUUUUUAGGCCCAUUUGUGCGCUUCAGCGCGGCCACACUAACCUUUCCUUCUUUUCUUACAAG .....(((((.(((.....(((((........((((((....((.......((((((((........))))))))........)).....))))))......))))).))))))))....................... (-23.82 = -25.13 + 1.32)
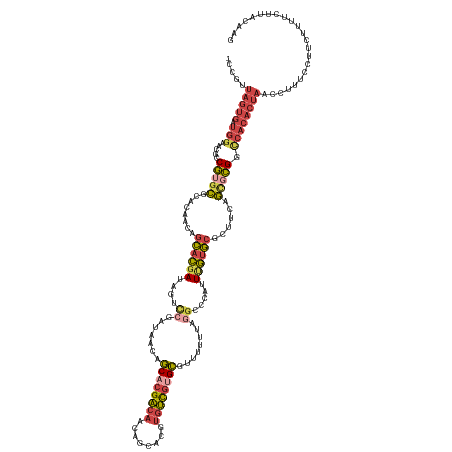
| Location | 15,329,738 – 15,329,876 |
|---|---|
| Length | 138 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Shannon entropy | 0.50717 |
| G+C content | 0.50432 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -27.50 |
| Energy contribution | -28.73 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
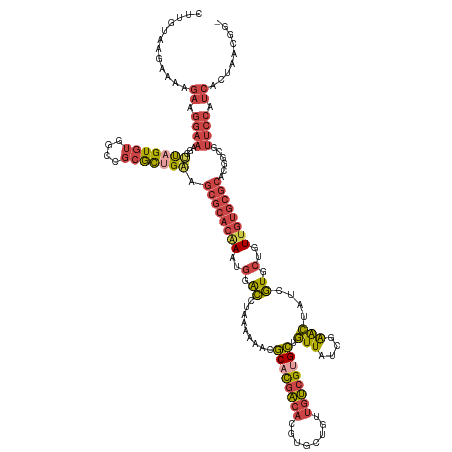
>dm3.chr2R 15329738 138 - 21146708 CUUGUAAGAAAAGAAGGAAAGGAUAGUGUGGCCGCGCUGAGGCGCACAAAUGGACCUAAAAAACGCACGACACGUGCUGUUGUCGUGCUGUUAUCCAACUAUCGUGCUGUUGUGCGCACGUCGUUCCAUCACUAACGG- ............((.((((..((((((.(((..((((....))))................(((((((((((........)))))))).)))..)))))))))((((......))))......)))).))........- ( -41.80, z-score = -0.29, R) >droSim1.chr2R 14042184 138 - 19596830 CUUGUAAGAAAAGAAGGAAAGGUUAGUGUGACCGCGCUGAAGCGCACAAAUGGACCUAAAAAACGCACGACACGUGCUGUUGUCGUGCUGUUAUCGAACUAUCGUGCUGUUGUGCGCACGGCCUUCCAUCACUAACGG- ............((.((((.(((((((((....))))))..((((((((..(.((......(((((((((((........)))))))).)))...((....)))).)..))))))))...))))))).))........- ( -43.90, z-score = -1.33, R) >droSec1.super_1 12845753 139 - 14215200 CUUGUAAGAAAAGAAGGAAAGGUUAGUGUGGCCGCGCUGAAGCGCACAAAUGAACCUAAAAAACGCACGACACGUGCUGUUGUCGUGCUGUUAUCGAACUAUCGUGCUGUUGUGCGCACGGCCUUCCAUCACUAACAGG .....................((((((((((..(.((((..((((((((...............((((((((........)))))))).((...(((....))).))..)))))))).)))))..))).)))))))... ( -45.70, z-score = -1.59, R) >droYak2.chr2R 7292471 138 + 21139217 CUUGUAAGAAAAGAAGGAAAGGUUAGUGUGGCCGCGCCGAAGCGCACGAAUGGGCCUAAAUAACGCACGACACGUGCUGUUGUCGUGCUGUUAUCGGACUAUCGUGCUGUUGUGCGCUCGCCGUUCCAUCACUAACGC- .....................(((((((((((.(((((((...((((((.(((.((...(((((((((((((........)))))))).))))).)).)))))))))..))).))))..))))......)))))))..- ( -51.20, z-score = -1.78, R) >droEre2.scaffold_4845 9528374 138 - 22589142 CUUGUAAGAAAAGAAGGAAAGGUUAGUGUGGCCGCACUGAAGCGCACGAAUGGGCCUAAAAAACGCUCGACACGUGCUGUUGUCGUGCUGUUAUCGGACUAUCGUGCUGUUGUGCGCACGCUGUUCCAUCACUAGCGG- ............((.((((....(((((((..(((((...(((((((((.(((.((.....(((((.(((((........))))).)).)))...)).))))))))).))))))))))))))))))).))........- ( -42.70, z-score = 0.28, R) >droAna3.scaffold_13266 17949152 112 + 19884421 ----CGAAAAGAAAAGAGGGAACCAGUGCUGCCAGAUGGGAGGAGGGCAGUCUGGCCAAAUACCACUUAU--UGUGGUACUAACGCGUUUUUGUCUGGGUUUUCUCAAAUU---CAAAUGG------------------ ----......(((..(((..((((.(.....)(((((..(((...(((......)))...((((((....--.))))))........)))..)))))))))..)))...))---)......------------------ ( -32.50, z-score = -1.13, R) >consensus CUUGUAAGAAAAGAAGGAAAGGUUAGUGUGGCCGCGCUGAAGCGCACAAAUGGACCUAAAAAACGCACGACACGUGCUGUUGUCGUGCUGUUAUCGAACUAUCGUGCUGUUGUGCGCACGGCGUUCCAUCACUAACGG_ ............((.((((...(((((((....))))))).((((((((..(.((.........((((((((........)))))))).(((....)))....)).)..))))))))......)))).))......... (-27.50 = -28.73 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:51 2011