| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,302,336 – 15,302,425 |
| Length | 89 |
| Max. P | 0.682512 |

| Location | 15,302,336 – 15,302,425 |
|---|---|
| Length | 89 |
| Sequences | 13 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Shannon entropy | 0.19710 |
| G+C content | 0.57724 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -28.35 |
| Energy contribution | -28.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
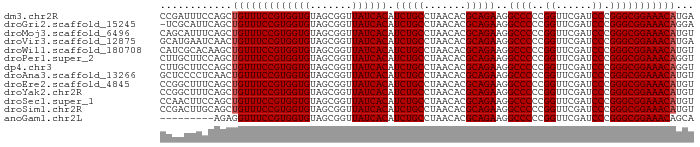
>dm3.chr2R 15302336 89 + 21146708 CCGAUUUCCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGA ............(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))... ( -28.60, z-score = -0.54, R) >droGri2.scaffold_15245 13573705 88 + 18325388 -UCGCAUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGA -..........((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).. ( -32.00, z-score = -1.37, R) >droMoj3.scaffold_6496 15765795 89 + 26866924 CAGCAUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU .(((......)))((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))).... ( -30.10, z-score = -1.29, R) >droVir3.scaffold_12875 9450944 89 - 20611582 GCAUGAAUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGA .......(((..(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))))) ( -28.90, z-score = -0.88, R) >droWil1.scaffold_180708 9060108 89 - 12563649 CAUCGCACAAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ....((....))(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))... ( -29.40, z-score = -0.85, R) >droPer1.super_2 6081051 89 - 9036312 CUUGCUUCCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGU ...........((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).. ( -32.00, z-score = -1.19, R) >dp4.chr3 17014135 89 - 19779522 CUUGCUUCCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGU ...........((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).. ( -32.00, z-score = -1.19, R) >droAna3.scaffold_13266 17922778 89 - 19884421 GCUCCCCUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ..........(((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))).)) ( -28.70, z-score = -0.88, R) >droEre2.scaffold_4845 9500914 89 + 22589142 CCGGCUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ..(((.....)))((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))).... ( -30.10, z-score = -0.57, R) >droYak2.chr2R 7265148 89 - 21139217 CCGGCUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ..(((.....)))((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))).... ( -30.10, z-score = -0.57, R) >droSec1.super_1 12815036 89 + 14215200 CCAACUUCCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ..........(((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))).)) ( -29.00, z-score = -0.96, R) >droSim1.chr2R 14014613 89 + 19596830 CCGACUUGCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ..........(((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))).)) ( -29.00, z-score = -0.29, R) >anoGam1.chr2L 19798871 80 - 48795086 ---------AGAGGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGCA ---------.(..((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))..). ( -28.90, z-score = -1.20, R) >consensus CCGGCUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGU ............(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))... (-28.35 = -28.43 + 0.08)


| Location | 15,302,336 – 15,302,425 |
|---|---|
| Length | 89 |
| Sequences | 13 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 91.93 |
| Shannon entropy | 0.19710 |
| G+C content | 0.57724 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
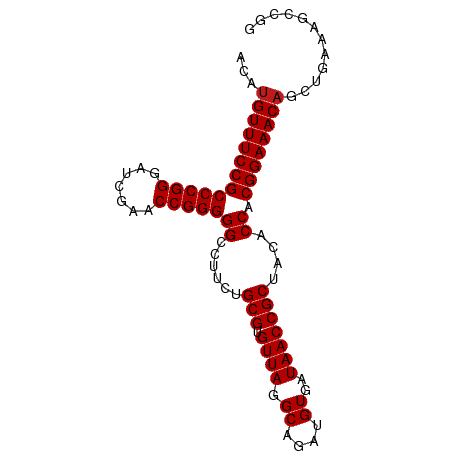
>dm3.chr2R 15302336 89 - 21146708 UCAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGGAAAUCGG ((.(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))....))...... ( -30.00, z-score = -0.69, R) >droGri2.scaffold_15245 13573705 88 - 18325388 UCCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAUGCGA- ..((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))..........- ( -31.90, z-score = -1.33, R) >droMoj3.scaffold_6496 15765795 89 - 26866924 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAUGCUG ....((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))(((......))). ( -31.30, z-score = -1.63, R) >droVir3.scaffold_12875 9450944 89 + 20611582 UCAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAUUCAUGC ((((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))..)))....... ( -29.90, z-score = -1.06, R) >droWil1.scaffold_180708 9060108 89 + 12563649 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUUGUGCGAUG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))((....)).... ( -31.40, z-score = -1.16, R) >droPer1.super_2 6081051 89 + 9036312 ACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGGAAGCAAG ..((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))........... ( -31.90, z-score = -1.23, R) >dp4.chr3 17014135 89 + 19779522 ACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGGAAGCAAG ..((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))........... ( -31.90, z-score = -1.23, R) >droAna3.scaffold_13266 17922778 89 + 19884421 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAGGGGAGC ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))............ ( -29.70, z-score = -0.26, R) >droEre2.scaffold_4845 9500914 89 - 22589142 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAGCCGG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))((.....))... ( -30.20, z-score = -0.71, R) >droYak2.chr2R 7265148 89 + 21139217 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAGCCGG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))((.....))... ( -30.20, z-score = -0.71, R) >droSec1.super_1 12815036 89 - 14215200 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGGAAGUUGG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))............ ( -29.70, z-score = -0.45, R) >droSim1.chr2R 14014613 89 - 19596830 ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGCAAGUCGG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))............ ( -29.70, z-score = -0.38, R) >anoGam1.chr2L 19798871 80 + 48795086 UGCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACCUCU--------- ....((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))....--------- ( -27.80, z-score = -0.92, R) >consensus ACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAGCCGG ...(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))............ (-29.33 = -29.41 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:46 2011