
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,299,429 – 15,299,555 |
| Length | 126 |
| Max. P | 0.802603 |

| Location | 15,299,429 – 15,299,521 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.47507 |
| G+C content | 0.34650 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.43 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
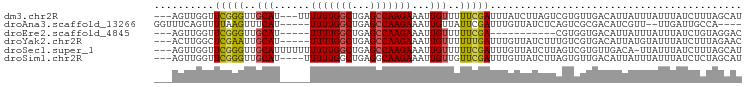
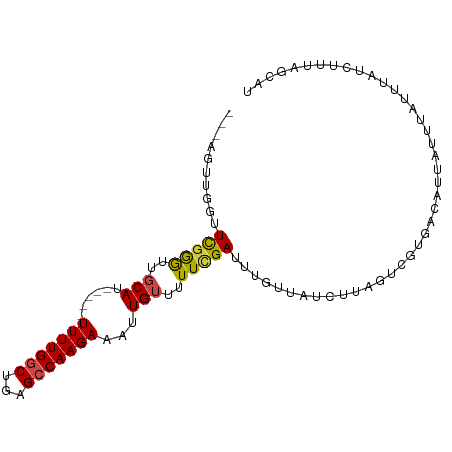
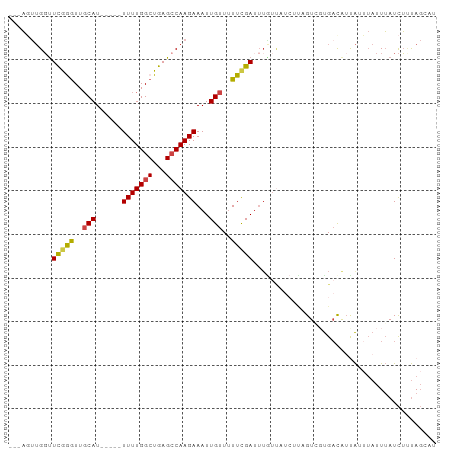
>dm3.chr2R 15299429 92 - 21146708 ---AGUUGGUUCGGGUUGCAU---UUUUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUAUCUUAGUCGUGUUGACAUUAUUUAUUUAUCUUUAGCAU ---.(((((.(((((..(((.---(((((((((...))))))))).)))..))))).........(((.....)))...............))))).. ( -21.60, z-score = -2.67, R) >droAna3.scaffold_13266 17920116 87 + 19884421 GGUUUCAGUUUUAAGUUUCAU-----UUUUGGCUGAGCCAAGAAAUUGUUAUUCGAUUUGUUAUCUCAGUCGCGACAUCGUU--UUGAUUGCCA---- (((.((((.............-----....(((((((..((.((((((.....)))))).))..)))))))(((....))).--))))..))).---- ( -22.20, z-score = -2.69, R) >droEre2.scaffold_4845 9497995 79 - 22589142 ---AGUUGGUUCGGGUUGCAU-----UUUUGGCUGAGCCAAGAAAUUGUUUUUCGA-----------CGUGGUGACAUUAUUUAUUUAUCUGUAGGAC ---.......(((((..((((-----(((((((...))))))))..)))..)))))-----------.(((....))).................... ( -17.00, z-score = -0.89, R) >droYak2.chr2R 7262236 90 + 21139217 ---ACUUGGCUCGAAUUGCAU-----UUUUGGCUGAGCCAAGAAAUUGUUUUUUGAUUUGUUAUCUUUGUCGUGACAUUAUGUAUUUAUCUUUAGAAC ---.(((((((((....((..-----.....))))))))))).....(((((..(((..((......(((....)))......))..)))...))))) ( -18.50, z-score = -1.64, R) >droSec1.super_1 12811981 94 - 14215200 ---AGUUGGUUCGGGUUGCAUUUUUUUUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUGUUAUCUUAGUCGUGUUGACA-UUAUUUAUCUUUAGCAU ---.(((((.(((((..(((....(((((((((...))))))))).)))..)))))..(((((.(........).)))))-..........))))).. ( -19.80, z-score = -1.80, R) >droSim1.chr2R 14011584 91 - 19596830 ---AGUUGGUUCGGGUUGCAU----UUUUUGGCUGAGGCAAGAAAUUGUUGUUCGAUUUGUUAUCUUAGUGUUGACAUUAUUUAUUUAUCUCUAGCAU ---.(((((.(((..(.(((.----(((((.((....)).))))).))).)..)))..(((((.(.....).)))))..............))))).. ( -18.60, z-score = -1.06, R) >consensus ___AGUUGGUUCGGGUUGCAU_____UUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUGUUAUCUUAGUCGUGACAUUAUUUAUUUAUCUUUAGCAU ..........(((((..(((......(((((((...)))))))...)))..))))).......................................... ( -9.54 = -9.43 + -0.11)
| Location | 15,299,458 – 15,299,555 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Shannon entropy | 0.39463 |
| G+C content | 0.35991 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.11 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
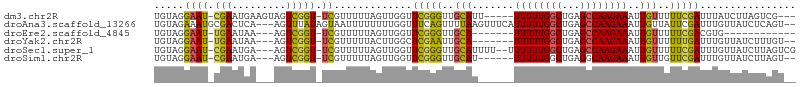
>dm3.chr2R 15299458 97 - 21146708 UGUAGGAAU-CGAAUGAAGUAGUCGGU-UCGUUUUUAGUUGGUUCGGGUUGCAUU-----UUUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUAUCUUAGUCG--- .....((((-(((.........)))))-))......((.(((.(((((..(((.(-----((((((((...))))))))).)))..))))).))).))......--- ( -24.40, z-score = -2.25, R) >droAna3.scaffold_13266 17920138 102 + 19884421 UGUAGAAAUGCGACUCA---AGUUUAUAGUAAUUUUUUUUGGUUUCAGUUUUAAGUUUCAUUUUUGGCUGAGCCAAGAAAUUGUUAUUCGAUUUGUUAUCUCAGU-- .(((....)))((..((---((((...((((((.((((((((((.(((((..((((...))))..)))))))))))))))..)))))).))))))...)).....-- ( -23.90, z-score = -2.93, R) >droEre2.scaffold_4845 9498022 83 - 22589142 UGUAGGAAU-UGAAUAA---AGUCGGU-UCGUUUUUAGUUGGUUCGGGUUGCA-------UUUUUGGCUGAGCCAAGAAAUUGUUUUUCGACGUG------------ .....((((-(((....---..)))))-)).............(((((..(((-------((((((((...))))))))..)))..)))))....------------ ( -19.40, z-score = -1.34, R) >droYak2.chr2R 7262264 93 + 21139217 UGUAGGAAU-UGAAUAA---AGUCGGU-UCGUUUUUACUUGGCUCGAAUUGCA-------UUUUUGGCUGAGCCAAGAAAUUGUUUUUUGAUUUGUUAUCUUUGU-- .((((((((-.((((..---.....))-)))))))))).(((((((((..(((-------((((((((...))))))))..)))..)))))...)))).......-- ( -19.80, z-score = -1.11, R) >droSec1.super_1 12812006 100 - 14215200 UGUAGGAAU-CGAAUGA---AGUCGGU-UCGUUUUUAGUUGGUUCGGGUUGCAUUUU--UUUUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUGUUAUCUUAGUCG .....((((-(((....---..)))))-)).............(((((..(((....--(((((((((...))))))))).)))..)))))................ ( -22.90, z-score = -1.59, R) >droSim1.chr2R 14011612 94 - 19596830 UGUAGGAAU-CGAAUGA---AGUCGGU-UCGUUUUUAGUUGGUUCGGGUUGCAU------UUUUUGGCUGAGGCAAGAAAUUGUUGUUCGAUUUGUUAUCUUAGU-- (((((((((-(((.(((---((.((..-.)).))))).)))))))...))))).------......((((((..((.((((((.....)))))).))..))))))-- ( -21.50, z-score = -1.64, R) >consensus UGUAGGAAU_CGAAUGA___AGUCGGU_UCGUUUUUAGUUGGUUCGGGUUGCAU______UUUUUGGCUGAGCCAAGAAAUUGUUUUUCGAUUUGUUAUCUUAGU__ .....................(((((...((.((((..(((((((((.(................).))))))))))))).))....)))))............... (-11.35 = -11.11 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:44 2011