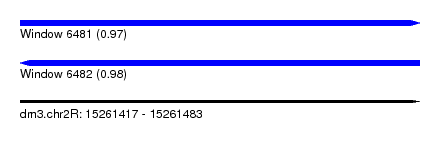
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,261,417 – 15,261,483 |
| Length | 66 |
| Max. P | 0.984973 |

| Location | 15,261,417 – 15,261,483 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 78.99 |
| Shannon entropy | 0.41596 |
| G+C content | 0.44753 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
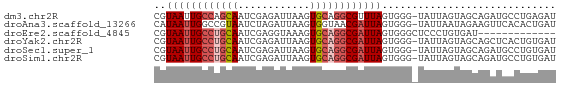
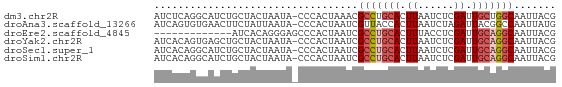
>dm3.chr2R 15261417 66 + 21146708 CGUAAUUGCCAGCAAUCGAGAUUAAGUGCAGGCGUUUAGUGGG-UAUUAGUAGCAGAUGCCUGAGAU ..(((.((((.(((............))).)))).)))..(((-((((.......)))))))..... ( -13.40, z-score = -0.09, R) >droAna3.scaffold_13266 17871485 66 - 19884421 CAUAAUUGGCCGUAAUCUAGAUUAAGUGGUAACGAUUAGUGGG-UAUUAAUAGAAGUUCACACUGAU ..(((((((((.((((....))))...)))..))))))(((..-(.((....)).)..)))...... ( -9.50, z-score = 0.23, R) >droEre2.scaffold_4845 9459301 54 + 22589142 CGUAAUUGCCUGCAAUCGAGGUAAAGUGCAGGCGAUUAGUGGGCUCCCUGUGAU------------- ..((((((((((((............))))))))))))..((....))......------------- ( -18.30, z-score = -2.07, R) >droYak2.chr2R 7224759 66 - 21139217 CGUAAUUGCCUGCAAUCGAGAUUAAGUGCAGGCGAUUAGUGGG-UAUUAGUAGCAGCUCACUGUGAU ..((((((((((((............))))))))))))(((((-(..........))))))...... ( -22.30, z-score = -2.72, R) >droSec1.super_1 12774593 66 + 14215200 CGUAAUUGCCUGCAAUCGAGAUUAAGUGCAGGCGAUUAGUGGG-UAUUAGUAGCAGAUGCCUGUGAU ..((((((((((((............))))))))))))(..((-((((.......))))))..)... ( -23.90, z-score = -3.81, R) >droSim1.chr2R 13973550 66 + 19596830 CGUAAUUGCCUGCAAUCGAGAUUAAGUGCAGGCGAUUAGUGGG-UAUUAGUAGCAGAUGCCUGUGAU ..((((((((((((............))))))))))))(..((-((((.......))))))..)... ( -23.90, z-score = -3.81, R) >consensus CGUAAUUGCCUGCAAUCGAGAUUAAGUGCAGGCGAUUAGUGGG_UAUUAGUAGCAGAUGCCUGUGAU ..((((((((((((............))))))))))))............................. (-11.99 = -12.85 + 0.86)
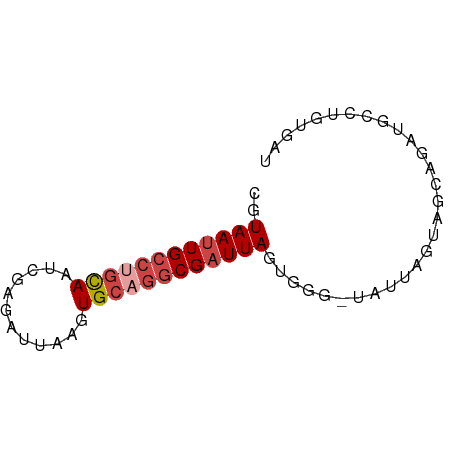
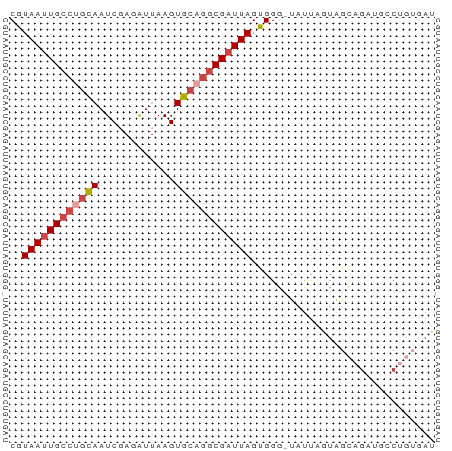
| Location | 15,261,417 – 15,261,483 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Shannon entropy | 0.41596 |
| G+C content | 0.44753 |
| Mean single sequence MFE | -12.40 |
| Consensus MFE | -8.77 |
| Energy contribution | -9.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
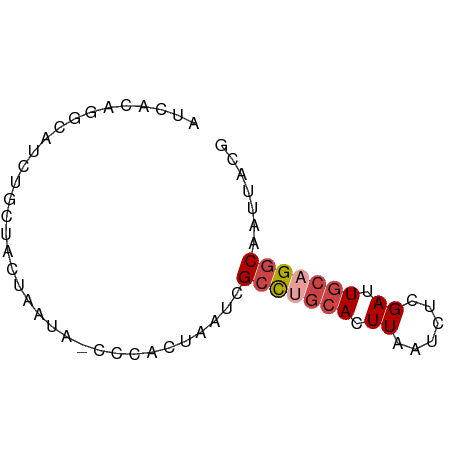
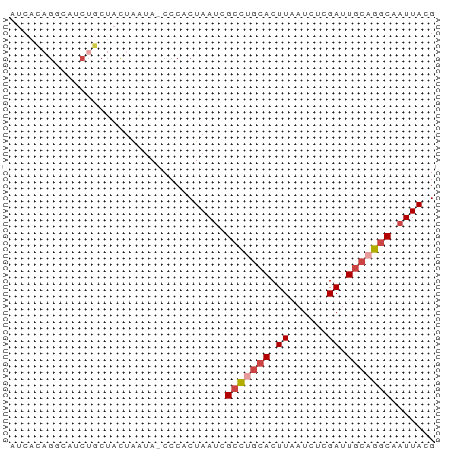
>dm3.chr2R 15261417 66 - 21146708 AUCUCAGGCAUCUGCUACUAAUA-CCCACUAAACGCCUGCACUUAAUCUCGAUUGCUGGCAAUUACG ......(((....))).......-..........(((.(((.((......)).))).)))....... ( -10.50, z-score = -1.56, R) >droAna3.scaffold_13266 17871485 66 + 19884421 AUCAGUGUGAACUUCUAUUAAUA-CCCACUAAUCGUUACCACUUAAUCUAGAUUACGGCCAAUUAUG ...((((..(((....((((...-.....)))).)))..))))........................ ( -2.70, z-score = 1.43, R) >droEre2.scaffold_4845 9459301 54 - 22589142 -------------AUCACAGGGAGCCCACUAAUCGCCUGCACUUUACCUCGAUUGCAGGCAAUUACG -------------......((....))..((((.(((((((.((......)).))))))).)))).. ( -15.50, z-score = -3.06, R) >droYak2.chr2R 7224759 66 + 21139217 AUCACAGUGAGCUGCUACUAAUA-CCCACUAAUCGCCUGCACUUAAUCUCGAUUGCAGGCAAUUACG ....(((....))).........-.....((((.(((((((.((......)).))))))).)))).. ( -14.90, z-score = -2.52, R) >droSec1.super_1 12774593 66 - 14215200 AUCACAGGCAUCUGCUACUAAUA-CCCACUAAUCGCCUGCACUUAAUCUCGAUUGCAGGCAAUUACG ......(((....))).......-.....((((.(((((((.((......)).))))))).)))).. ( -15.40, z-score = -3.54, R) >droSim1.chr2R 13973550 66 - 19596830 AUCACAGGCAUCUGCUACUAAUA-CCCACUAAUCGCCUGCACUUAAUCUCGAUUGCAGGCAAUUACG ......(((....))).......-.....((((.(((((((.((......)).))))))).)))).. ( -15.40, z-score = -3.54, R) >consensus AUCACAGGCAUCUGCUACUAAUA_CCCACUAAUCGCCUGCACUUAAUCUCGAUUGCAGGCAAUUACG ..................................(((((((.((......)).)))))))....... ( -8.77 = -9.47 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:43 2011