| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,225,701 – 15,225,769 |
| Length | 68 |
| Max. P | 0.956651 |

| Location | 15,225,701 – 15,225,769 |
|---|---|
| Length | 68 |
| Sequences | 10 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.60493 |
| G+C content | 0.57017 |
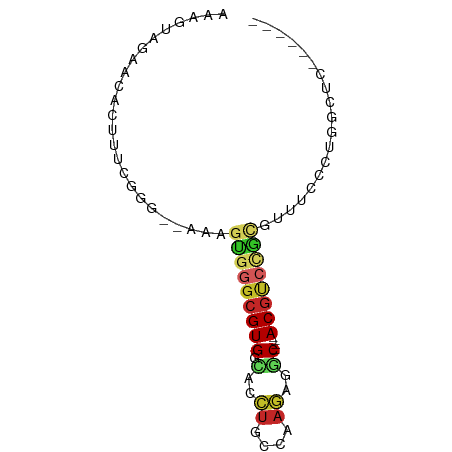
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.32 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
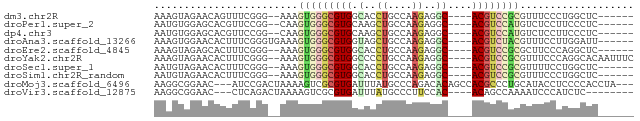
>dm3.chr2R 15225701 68 + 21146708 AAAGUAGAACAGUUUCGGG--AAAGUGGGCGUGGCACCUGCCAAGAGGC----ACGUCCGCGUUUCCCUGGCUC------ ..........((((..(((--((((((((((((.(..((....))..))----)))))))).)))))).)))).------ ( -28.00, z-score = -2.27, R) >droPer1.super_2 7585225 68 + 9036312 AAUGUGGAGCACGUUCCGG--CAAGUGGGCGUGCAAGCUGCCAAGAGGC----ACGUCCAUGUCUCCUUCCCUC------ .....((((.((((....)--)..((((((((((...((....))..))----)))))))))))))).......------ ( -25.60, z-score = -1.64, R) >dp4.chr3 7382923 68 + 19779522 AAUGUGGAGCACGUUCCGG--CAAGUGGGCGUGCAAGCUGCCAAGAGGC----ACGUCCAUGUCUCCUUCCCUC------ .....((((.((((....)--)..((((((((((...((....))..))----)))))))))))))).......------ ( -25.60, z-score = -1.64, R) >droAna3.scaffold_13266 17830677 70 - 19884421 AAAGUGGAACACUUUCGGGUGAAAGUGGGCGUGGUAGCUGCCAAGAGGC----ACGUCUACGUUUCCUUGGAUU------ .....(((((((((((....))))))).((((((..((((((....)))----).)))))))))))).......------ ( -23.70, z-score = -1.74, R) >droEre2.scaffold_4845 9423241 68 + 22589142 AAAGUAGAGCACUUUCGGG--AAAGUGGGCGUGGCACCUGCCAAGAGGC----ACGUCCGCGCUUCCCAGGCUC------ ......((((.((...(((--((.(((((((((.(..((....))..))----))))))))..)))))))))))------ ( -30.90, z-score = -2.64, R) >droYak2.chr2R 7188198 74 - 21139217 AAAGUAGAACACUUUCGGG--AAAGUGGGCGUGGCCCCUGCCAAGAGGC----ACGUCCGCGUUUCCCAGGCACAAUUUC (((((.....))))).(((--((((((((((((.((.((....)).)))----)))))))).))))))............ ( -29.90, z-score = -2.80, R) >droSec1.super_1 12739267 68 + 14215200 AAUGUAGAACACUUUCGGG--AAAGUGGGCGUGGCACCUGCCAAGAGGC----ACGUCCGCGUUUUCCUGGCUC------ ..............(((((--((.(((((((((.(..((....))..))----))))))))...)))))))...------ ( -25.90, z-score = -1.73, R) >droSim1.chr2R_random 2643221 68 + 2996586 AAUGUAGAACACUUUCGGG--AAAGUGGGCGUGGCACCUGCCAAGAGGC----ACGUCCGCGUUUCCCUGGCUC------ ................(((--((((((((((((.(..((....))..))----)))))))).))))))......------ ( -27.00, z-score = -1.85, R) >droMoj3.scaffold_6496 14938511 74 - 26866924 AAGGCGGAAC---AUCCGACUAAAAGUCGCGUGAUUUAUGCCCAGACACAGCCACGCCCUGCAUACCUCCCCACCUA--- .(((.((..(---((.((((.....)))).)))...(((((...................))))).....)).))).--- ( -15.21, z-score = -1.24, R) >droVir3.scaffold_12875 2331281 65 - 20611582 AAGGCGGAAC---CUCAGACUAAAAGUCGCGUGAUUUAUGCCCUUCCAC----ACAGCCAAAAUCCCAUCUC-------- ..(((((((.---....(((.....)))(((((...)))))..))))..----...))).............-------- ( -11.40, z-score = -0.99, R) >consensus AAAGUAGAACACUUUCGGG__AAAGUGGGCGUGGCACCUGCCAAGAGGC____ACGUCCGCGUUUCCCUGGCUC______ ........................((((((((.......(((....)))....))))))))................... (-10.60 = -11.32 + 0.72)
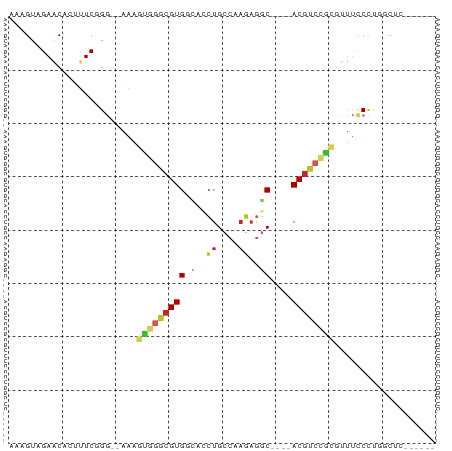
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:40 2011