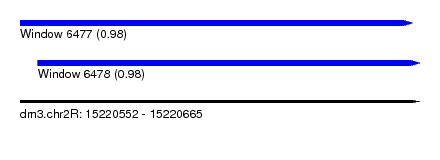
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,220,552 – 15,220,665 |
| Length | 113 |
| Max. P | 0.980458 |

| Location | 15,220,552 – 15,220,663 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.97 |
| Shannon entropy | 0.55626 |
| G+C content | 0.40797 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
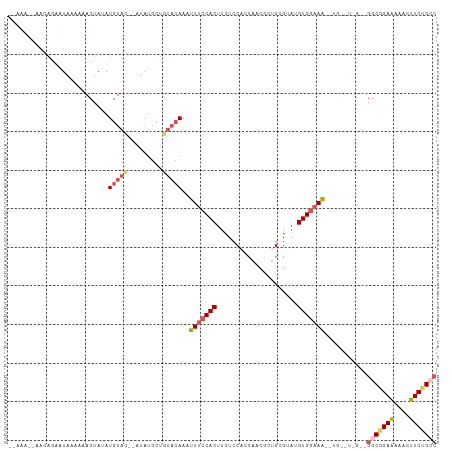
>dm3.chr2R 15220552 111 + 21146708 UAAAAACAACAGAAUAAAAAACUAAAUGUAUAUGUAUGCUGUACAAAUUUCCACUUCUACUUCAUCGCUGCGUAUGUGGAAAACUGAGUAGAAGGCGGAAAAACUUCUGCC .........(((((............((((((.(....)))))))..(((((.(((((((((.....(..(....)..)......)))))))))..)))))...))))).. ( -26.60, z-score = -3.61, R) >droEre2.scaffold_4845 9418237 88 + 22589142 ----------AGAAUAAGAAACUAUAUGUAC-------------AAACUCCCACUUUUCCACCAUCCCUGUGUACGUGGCAGGCUGGUUGGUGGGCGGAGAAACUUCCGCC ----------.....................-------------......(((((.....((((..(((((.......))))).)))).)))))((((((....)))))). ( -23.70, z-score = -0.39, R) >droYak2.chr2R 7182692 109 - 21139217 AAAAAGAAACGGACUAAAAAACUACAUGUAC--AUAUGCCGUACAAAUUUCCACUCCUCCCACCACACUGUGCAUGUGAAAAAGUGAGUGGAAGGCGGAAAAACUUCGGCC ..........................(((((--.......)))))...((((((((((.....((((.......))))....)).))))))))(((.(((....))).))) ( -25.00, z-score = -1.60, R) >droSec1.super_1 12734132 92 + 14215200 -------UAAAAAAUAAAAAACUAUAUGUACCAAUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA-----------GGCGGAAAAACUUCUGG- -------...................(((((.........)))))..(((((.(((.(((((..(((...)))..))))).)-----------)).))))).........- ( -18.50, z-score = -2.52, R) >droSim1.chr2R 13931285 98 + 19596830 UGAAAACAACAGAAUAAAAAACUAUAUGUAC--AUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA-----------GGCGGAAAAACUUCUGGC .........(((((............(((((--(.....))))))..(((((.(((.(((((..(((...)))..))))).)-----------)).)))))...))))).. ( -25.80, z-score = -3.95, R) >consensus __AAA__AACAGAAUAAAAAACUAUAUGUAC__AUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA__UG__U_G__GGCGGAAAAACUUCUGCC ...............................................(((((((.....................)))))))...........(((((((....))))))) (-10.12 = -10.72 + 0.60)

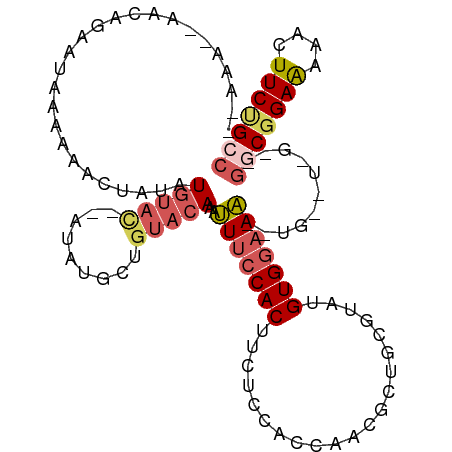
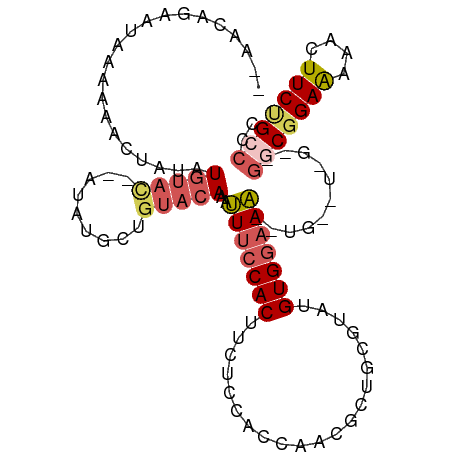
| Location | 15,220,557 – 15,220,665 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.53685 |
| G+C content | 0.42898 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -10.76 |
| Energy contribution | -11.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15220557 108 + 21146708 ACAACAGAAUAAAAAACUAAAUGUAUAUGUAUGCUGUACAAAUUUCCACUUCUACUUCAUCGCUGCGUAUGUGGAAAACUGAGUAGAAGGCGGAAAAACUUCUGCCCC ....(((((............((((((.(....)))))))..(((((.(((((((((.....(..(....)..)......)))))))))..)))))...))))).... ( -26.60, z-score = -3.27, R) >droEre2.scaffold_4845 9418237 90 + 22589142 -----AGAAUAAGAAACUAUAUGUAC-------------AAACUCCCACUUUUCCACCAUCCCUGUGUACGUGGCAGGCUGGUUGGUGGGCGGAGAAACUUCCGCCCC -----.....................-------------..............((((((..(((((.......))))).))).))).((((((((....)))))))). ( -26.00, z-score = -0.85, R) >droYak2.chr2R 7182697 106 - 21139217 GAAACGGACUAAAAAACUACAUGUAC--AUAUGCCGUACAAAUUUCCACUCCUCCCACCACACUGUGCAUGUGAAAAAGUGAGUGGAAGGCGGAAAAACUUCGGCCCU .....................(((((--.......)))))...((((((((((.....((((.......))))....)).))))))))(((.(((....))).))).. ( -25.80, z-score = -1.78, R) >droSec1.super_1 12734137 87 + 14215200 -------AAUAAAAAACUAUAUGUACCAAUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA-----------GGCGGAAAAACUUCUGG--- -------..............(((((.........)))))..(((((.(((.(((((..(((...)))..))))).)-----------)).))))).........--- ( -18.50, z-score = -2.22, R) >droSim1.chr2R 13931290 95 + 19596830 ACAACAGAAUAAAAAACUAUAUGUAC--AUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA-----------GGCGGAAAAACUUCUGGCCC ....(((((............(((((--(.....))))))..(((((.(((.(((((..(((...)))..))))).)-----------)).)))))...))))).... ( -25.80, z-score = -3.74, R) >consensus __AACAGAAUAAAAAACUAUAUGUAC__AUAUGCUGUACAAAUUUCCACUUCUCCACCAACGCUGCGUAUGUGGAAA__UG__U_G__GGCGGAAAAACUUCUGCCCC ..........................................(((((((.....................)))))))...........(((((((....))))))).. (-10.76 = -11.36 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:39 2011