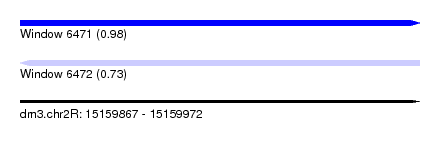
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,159,867 – 15,159,972 |
| Length | 105 |
| Max. P | 0.983751 |

| Location | 15,159,867 – 15,159,972 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Shannon entropy | 0.40608 |
| G+C content | 0.41890 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -19.50 |
| Energy contribution | -22.47 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
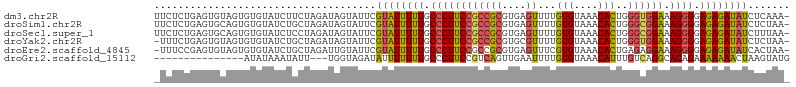
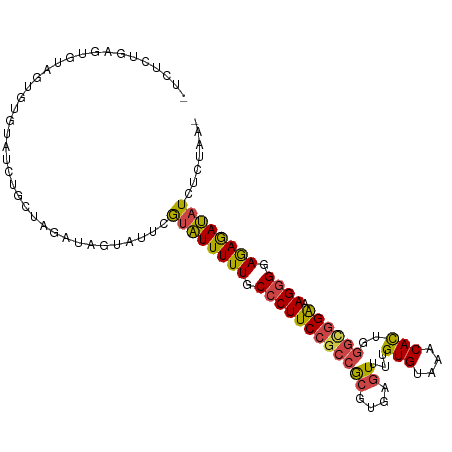
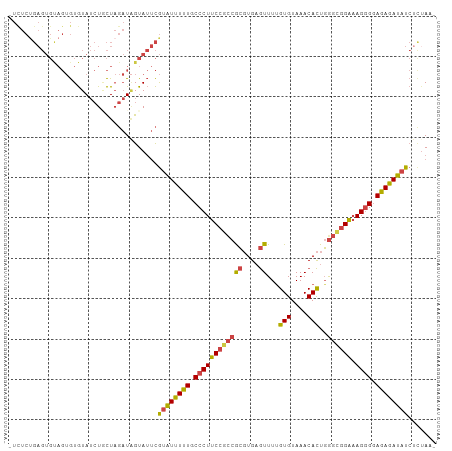
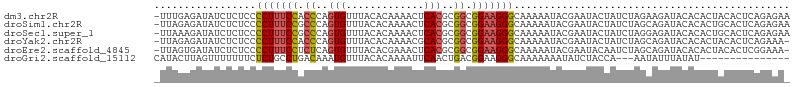
>dm3.chr2R 15159867 105 + 21146708 UUCUCUGAGUGUAGUGUGUAUCUUCUAGAUAGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGAGUUUUGUGUAAACACUGGGUGGAAAGGGGAGAGAUAUCUCAAA- .....((((.(((.(((.((.....)).))).)))..((((((((.((((((((((((....))...(((....)))..))))).))))).))))))))))))..- ( -32.90, z-score = -2.87, R) >droSim1.chr2R 13870311 105 + 19596830 UUCUCUGAGUGCAGUGUGUAUCUGCUAGAUAGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGAGUUUUGUGUAAACACUGGGCGGAAAGGGGAGAGAUAUCUCUAA- ......(((.((((.......))))............((((((((.((((((((((((....))...(((....)))..))))).))))).)))))))))))...- ( -36.20, z-score = -2.90, R) >droSec1.super_1 12674483 105 + 14215200 UUCUCUGAGUGCAGUGUGUAUCUCCUAGAUAGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGAGUUUUGUGUAAACACUGGGCGGAAAGGGGAGAGAUAUCUUUAA- ......((((((.((.((.......)).)).))))))((((((((.((((((((((((....))...(((....)))..))))).))))).))))))))......- ( -35.70, z-score = -3.11, R) >droYak2.chr2R 7120350 104 - 21139217 -UUUCUGAGUGUAGUGUGUAUCUGCUAGAUAGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGCGUUUUGUGUAAACACUGGGUGGAAAGGGGAGAGAUAUCUCUAA- -.....(((.((((.......))))............((((((((.((((((((((.((.(.((((.....)))).)))))))).))))).)))))))))))...- ( -30.20, z-score = -1.75, R) >droEre2.scaffold_4845 9363678 104 + 22589142 -UUUCCGAGUGUAGUGUGUAUCUGCUAGAUUGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGAGUUUCGUGUAAACACUGAGAGGAAAGGGGAGAGAUAUCACUAA- -....((((((((((.(((....)).).))))))))))(((((((.(((((((...((....))((((((....))).))).)).))))).))))))).......- ( -27.50, z-score = -1.10, R) >droGri2.scaffold_15112 3637651 88 + 5172618 ---------------AUAUAAAUAUU---UGGUAGAUAUUUUUUUGCCCUUCCGUCAGUUGAAUUUUGUGUAAACAUUUGUCAGGCAGAGAAAAAAACUAAGUAUG ---------------......(((((---((((.....((((((((((((......)).(((....(((....)))....)))))))))))))...))))))))). ( -18.10, z-score = -2.17, R) >consensus _UCUCUGAGUGUAGUGUGUAUCUGCUAGAUAGUAUUCGUAUUUUUGCCCUUCCGCCGCGUGAGUUUUGUGUAAACACUGGGCGGAAAGGGGAGAGAUAUCUCUAA_ ......((((((.((.((.......)).)).))))))((((((((.((((((((((((....))...(((....)))..)))))).)))).))))))))....... (-19.50 = -22.47 + 2.96)
| Location | 15,159,867 – 15,159,972 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Shannon entropy | 0.40608 |
| G+C content | 0.41890 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.19 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.729881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
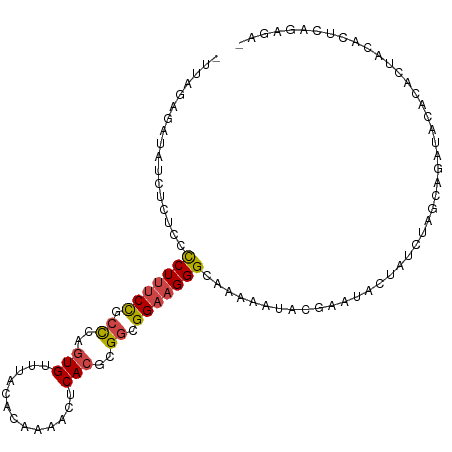
>dm3.chr2R 15159867 105 - 21146708 -UUUGAGAUAUCUCUCCCCUUUCCACCCAGUGUUUACACAAAACUCACGCGGCGGAAGGGCAAAAAUACGAAUACUAUCUAGAAGAUACACACUACACUCAGAGAA -((((((..........(((((((.((..(((.............)))..)).)))))))...............((((.....)))).........))))))... ( -21.32, z-score = -1.93, R) >droSim1.chr2R 13870311 105 - 19596830 -UUAGAGAUAUCUCUCCCCUUUCCGCCCAGUGUUUACACAAAACUCACGCGGCGGAAGGGCAAAAAUACGAAUACUAUCUAGCAGAUACACACUGCACUCAGAGAA -.........(((((..((((((((((..(((.............)))..)))))))))).....................((((.......))))....))))). ( -29.52, z-score = -3.84, R) >droSec1.super_1 12674483 105 - 14215200 -UUAAAGAUAUCUCUCCCCUUUCCGCCCAGUGUUUACACAAAACUCACGCGGCGGAAGGGCAAAAAUACGAAUACUAUCUAGGAGAUACACACUGCACUCAGAGAA -.......(((((((..((((((((((..(((.............)))..)))))))))).........((......))..)))))))....(((....))).... ( -26.42, z-score = -2.75, R) >droYak2.chr2R 7120350 104 + 21139217 -UUAGAGAUAUCUCUCCCCUUUCCACCCAGUGUUUACACAAAACGCACGCGGCGGAAGGGCAAAAAUACGAAUACUAUCUAGCAGAUACACACUACACUCAGAAA- -...(((..........(((((((.(((.((((((.....))))))..).)).)))))))...............((((.....)))).........))).....- ( -21.50, z-score = -2.30, R) >droEre2.scaffold_4845 9363678 104 - 22589142 -UUAGUGAUAUCUCUCCCCUUUCCUCUCAGUGUUUACACGAAACUCACGCGGCGGAAGGGCAAAAAUACGAAUACAAUCUAGCAGAUACACACUACACUCGGAAA- -.(((((.(((((((..(((((((.((..(((.............)))..)).))))))).........((......)).)).)))))..)))))..........- ( -19.02, z-score = -0.51, R) >droGri2.scaffold_15112 3637651 88 - 5172618 CAUACUUAGUUUUUUUCUCUGCCUGACAAAUGUUUACACAAAAUUCAACUGACGGAAGGGCAAAAAAAUAUCUACCA---AAUAUUUAUAU--------------- ........((((((((((((..(((.((..((.............))..)).))).)))).))))))))........---...........--------------- ( -9.22, z-score = -0.12, R) >consensus _UUAGAGAUAUCUCUCCCCUUUCCGCCCAGUGUUUACACAAAACUCACGCGGCGGAAGGGCAAAAAUACGAAUACUAUCUAGCAGAUACACACUACACUCAGAGA_ .................(((((((.((..(((.............)))..)).))))))).............................................. (-10.94 = -11.19 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:34 2011