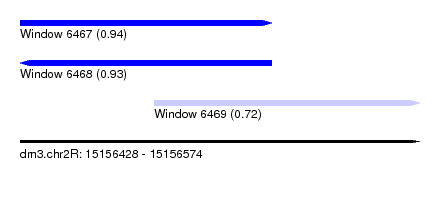
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,156,428 – 15,156,574 |
| Length | 146 |
| Max. P | 0.938693 |

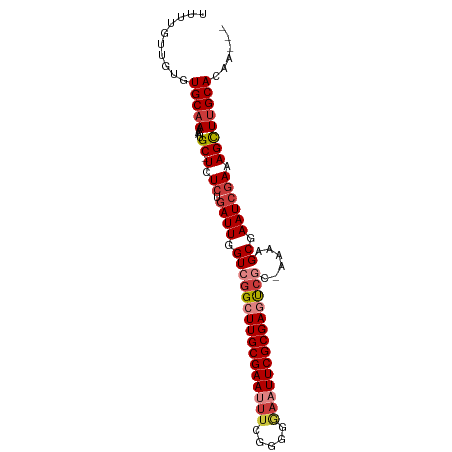
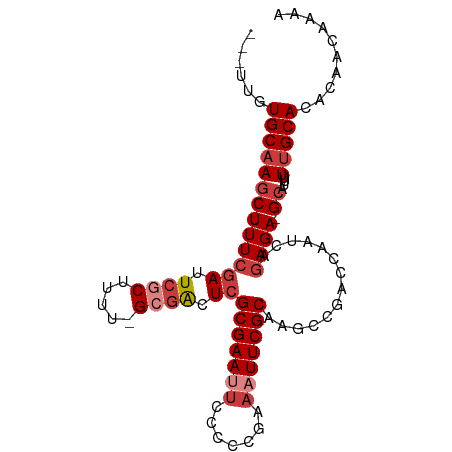
| Location | 15,156,428 – 15,156,520 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.23403 |
| G+C content | 0.45587 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -22.07 |
| Energy contribution | -23.56 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
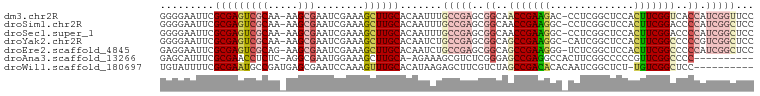
>dm3.chr2R 15156428 92 + 21146708 UUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA--- ........(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).--- ( -37.10, z-score = -3.96, R) >droSim1.chr2R 13867416 92 + 19596830 UUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA--- ........(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).--- ( -37.10, z-score = -3.96, R) >droSec1.super_1 12671030 92 + 14215200 UUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA--- ........(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).--- ( -37.10, z-score = -3.96, R) >droYak2.chr2R 7116800 92 - 21139217 UUUUGUUGUUUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC-AAAAGCGAAUCGAAAGCUUGCACAA--- ..........(((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).))))))))...--- ( -32.50, z-score = -2.47, R) >droEre2.scaffold_4845 9360166 92 + 22589142 UUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGCUUGCGAAUUUCGGAGGAAUUCGCGAGUCGC-AGAAGCGAAUCGAAAGCUUGCACAA--- ........(((((((....((-(.((.((((.((((((((((((((((.....)))))))))))))).-....)).)))))).)))))))))).--- ( -37.10, z-score = -3.46, R) >droAna3.scaffold_13266 8474750 95 - 19884421 UUUUGUUGUGUGCAAAAAUGC-UCUCUGAUUGGUCGGAUUGCGAAUUUCGGAGCAUUUCGCGAACCUC-UCAGGCGAAUGGAAAGCUUGCAAGAAAG .(((.(((((.((...(((((-(((..((((.((......)).))))..))))))))((((.......-....)))).......)).))))).))). ( -24.70, z-score = -0.00, R) >droWil1.scaffold_180697 1208313 97 - 4168966 UUUUGUUAUUUGCCAAUUUGCCUCUCUGAUUGGUCGGAUUGCGAAUUUCGUGUAUUUUCGCGAAUGCCGAUGAGCGAAUCCAAAGUUUGCACAUAAG .(((((.(((((((((((.........)))))).))))).))))).((((((......)))))).....(((.((((((.....)))))).)))... ( -24.10, z-score = -0.99, R) >consensus UUUUGUUGUGUGCAAAAAUGC_UCUCUGAUUGGUCGGCUUGCGAAUUUCGGGGGAAUUCGCGAGUCGC_AAAAGCGAAUCGAAAGCUUGCACAA___ ((((((.....)))))).(((..((..((((.((((((((((((((((.....))))))))))))))......)).))))...))...)))...... (-22.07 = -23.56 + 1.49)

| Location | 15,156,428 – 15,156,520 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.23403 |
| G+C content | 0.45587 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.63 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15156428 92 - 21146708 ---UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAAAA ---.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))........ ( -30.70, z-score = -4.63, R) >droSim1.chr2R 13867416 92 - 19596830 ---UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAAAA ---.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))........ ( -30.70, z-score = -4.63, R) >droSec1.super_1 12671030 92 - 14215200 ---UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAAAA ---.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))........ ( -30.70, z-score = -4.63, R) >droYak2.chr2R 7116800 92 + 21139217 ---UUGUGCAAGCUUUCGAUUCGCUUUU-GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCAAACAACAAAA ---...(((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....))))).......... ( -26.70, z-score = -3.04, R) >droEre2.scaffold_4845 9360166 92 - 22589142 ---UUGUGCAAGCUUUCGAUUCGCUUCU-GCGACUCGCGAAUUCCUCCGAAAUUCGCAAGCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAAAA ---.(((((((((((((((.((((....-)))).))(((((((.......)))))))..............))))-))....)))))))........ ( -30.70, z-score = -4.04, R) >droAna3.scaffold_13266 8474750 95 + 19884421 CUUUCUUGCAAGCUUUCCAUUCGCCUGA-GAGGUUCGCGAAAUGCUCCGAAAUUCGCAAUCCGACCAAUCAGAGA-GCAUUUUUGCACACAACAAAA (((((..((.............))..))-)))....((((((((((((((...(((.....)))....)).).))-))).))))))........... ( -18.82, z-score = -0.08, R) >droWil1.scaffold_180697 1208313 97 + 4168966 CUUAUGUGCAAACUUUGGAUUCGCUCAUCGGCAUUCGCGAAAAUACACGAAAUUCGCAAUCCGACCAAUCAGAGAGGCAAAUUGGCAAAUAACAAAA ......(((...(((((((.....)).((((.(((.(((((...........)))))))))))).....)))))..)))..(((........))).. ( -17.00, z-score = 0.16, R) >consensus ___UUGUGCAAGCUUUCGAUUCGCUUUU_GCGACUCGCGAAUUCCCCCGAAAUUCGCAAGCCGACCAAUCAGAGA_GCAUUUUUGCACACAACAAAA ......(((((((((((((.((((.....)))).))(((((((.......)))))))..............)))).))....))))).......... (-15.32 = -16.63 + 1.31)
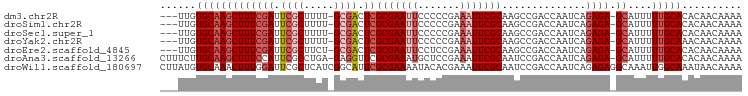
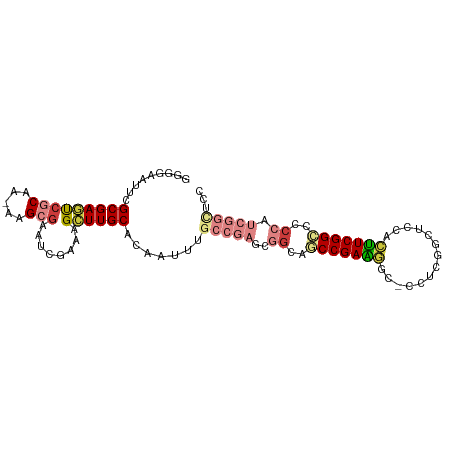
| Location | 15,156,477 – 15,156,574 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Shannon entropy | 0.52615 |
| G+C content | 0.59915 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15156477 97 + 21146708 GGGGAAUUCGCGAGUCGCAA-AAGCGAAUCGAAAGCUUGCACAAUUUGCCGAGCGGCAACCGAAGAC-CCUCGGCUCCACUUCGGUCACCAUCGGUUCC ..(((....(((((((((..-..))))..(....)))))).......(((((..((..(((((((..-...........)))))))..)).)))))))) ( -31.22, z-score = -0.63, R) >droSim1.chr2R 13867465 97 + 19596830 GGGGAAUUCGCGAGUCGCAA-AAGCGAAUCGAAAGCUUGCACAAUUUGCCGAGCGGCAACCGAAGGC-CCUCGGCUCCACUUCGGACCCCAUCGGCUCC ((((.....(((((((((..-..))))..(....)))))).......((((((.(((........))-)))))))(((.....)))))))......... ( -37.80, z-score = -1.76, R) >droSec1.super_1 12671079 97 + 14215200 GGGGAAUUCGCGAGUCGCAA-AAGCGAAUCGAAAGCUUGCACAAUUUGCCGAGCGGCAACCGAAGGC-CCUCGGCUCCACUUCGGACCCCAUCGGCUCC ((((.....(((((((((..-..))))..(....)))))).......((((((.(((........))-)))))))(((.....)))))))......... ( -37.80, z-score = -1.76, R) >droYak2.chr2R 7116849 97 - 21139217 GGGGAAUUCGCGAGUCGCAA-AAGCGAAUCGAAAGCUUGCACAAUCUGCCGAGCGGCAGCCGAAGGC-CAUCGGCUCCACUUCGGCCCCCGUCGGCUCC ((((.....(((((((((..-..))))..(....)))))).......((((((.((.((((((....-..)))))))).).)))))))))......... ( -40.90, z-score = -1.66, R) >droEre2.scaffold_4845 9360215 97 + 22589142 GAGGAAUUCGCGAGUCGCAG-AAGCGAAUCGAAAGCUUGCACAAUCUGCCGAGCGGCAGCCGAAGGG-UCUCGGCUCCACUUCGGCCCCCAUCGGCUCC ..(((....(((((((((..-..))))..(....)))))).......(((((..((..(((((((((-........)).)))))))..)).)))))))) ( -39.60, z-score = -1.43, R) >droAna3.scaffold_13266 8474799 87 - 19884421 GAGCAUUUCGCGAACCUCUC-AGGCGAAUGGAAAGCUUGCA-AGAAAGCGUCUCGGGAGCCGAGGCCACUUCGGCCCCCGUUCGGCCCC---------- ((((..(((((.........-..)))))......((((...-...))))).)))(((.(((((((((.....)))).....))))))))---------- ( -31.20, z-score = -0.15, R) >droWil1.scaffold_180697 1208363 88 - 4168966 UGUAUUUUCGCGAAUGCCGAUGAGCGAAUCCAAAGUUUGCACAUAAGAGCUUCGUCUAGCCGACACACAAUCGGCUCU-UGUCGGCUCC---------- .........(((((.((..(((.((((((.....)))))).)))....)))))))..((((((((.............-))))))))..---------- ( -24.12, z-score = -0.99, R) >consensus GGGGAAUUCGCGAGUCGCAA_AAGCGAAUCGAAAGCUUGCACAAUUUGCCGAGCGGCAGCCGAAGGC_CCUCGGCUCCACUUCGGCCCCCAUCGGCUCC ((((.....(((((((((.....)))........))))))......((((....))))(((((((..............)))))))))))......... (-18.85 = -19.88 + 1.03)
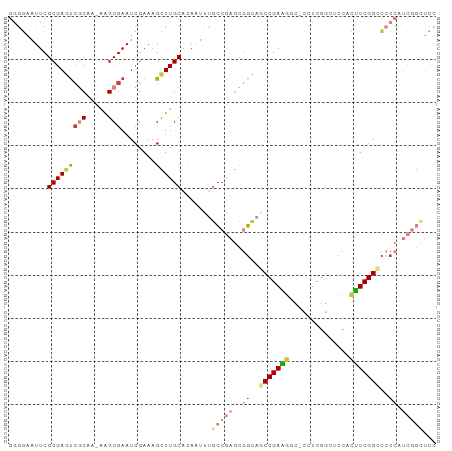
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:31 2011