| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,268,936 – 3,269,026 |
| Length | 90 |
| Max. P | 0.790360 |

| Location | 3,268,936 – 3,269,026 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.66 |
| Shannon entropy | 0.73552 |
| G+C content | 0.38311 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -7.75 |
| Energy contribution | -8.06 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
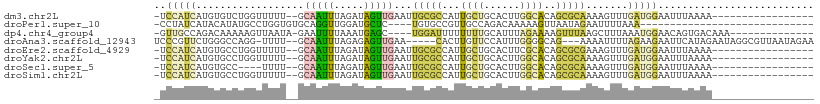
>dm3.chr2L 3268936 90 - 23011544 -UCCAUCAUGUGUCUGGUUUUU--GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAA----------------- -(((((((((((.(((......--(((((((((....)))))))))((((..........)))).)))))))......)))))))........----------------- ( -28.40, z-score = -2.40, R) >droPer1.super_10 324421 76 - 3432795 -CCUAUCAUACAUAUGCCUGGUGUGCAGGUUGGAUGCUC----UGUGCCGUUGCCAGACAAAAAGUUAAUAGAAUUUUAAA----------------------------- -.((((...((...((.((((((..(((((.....)).)----))..)....))))).))....))..)))).........----------------------------- ( -15.60, z-score = 0.06, R) >dp4.chr4_group4 1327630 90 - 6586962 -GUUGCCAGACAAAAAGUUAAUA-GAAUUUUAAAUGAGC----UGGAUUUUUUUUGCAUUUAGAAAAGUUUAAGCUUUAAAUGGAACAGUGACAAA-------------- -(((.(((.....((((((....-.))))))....((((----(((((((((((.......)))))))))).)))))....)))))).........-------------- ( -14.20, z-score = -0.02, R) >droAna3.scaffold_12943 23356 99 - 5039921 UCCCGUUCUGGGCCAGG-UUUU--GCAAUUUAGAGAGUUGAA-----CACUUGUUCCAUUUGGGGCAG---AAAAUUUUAGAAGAAUUCAUAGAAUAGGCGUUAAUAGAA .((((...(((..((((-(.((--.((((((...))))))))-----.)))))..)))..))))((..---...((((((((.....)).))))))..)).......... ( -20.60, z-score = -0.06, R) >droEre2.scaffold_4929 3300360 90 - 26641161 -UCCAUCAUGUGCCUGGUUUUU--GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUCGCACAGCGCGAAAGUUUGAUGGAAUUUAAAA----------------- -(((((((.((((.((((....--(((((((((....))))))))))))).....))))(((((.....)))))....)))))))........----------------- ( -28.80, z-score = -2.57, R) >droYak2.chr2L 3252050 90 - 22324452 -UCCAUCAUGUGCCUGGUUUUU--GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAA----------------- -(((((((..((((((......--(((((((((....)))))))))((((..........)))).))).)))......)))))))........----------------- ( -29.30, z-score = -2.45, R) >droSec1.super_5 1411221 86 - 5866729 -UCCAUCAUGUGCC----UUUU--GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAA----------------- -((((((((((((.----....--(((((((((....)))))))))((((..........))))...)))))......)))))))........----------------- ( -27.20, z-score = -2.49, R) >droSim1.chr2L 3205778 90 - 22036055 -UCCAUCAUGUGCCUGGUUUUU--GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAA----------------- -(((((((..((((((......--(((((((((....)))))))))((((..........)))).))).)))......)))))))........----------------- ( -29.30, z-score = -2.45, R) >consensus _UCCAUCAUGUGCCUGGUUUUU__GCAAUUUAGAUAGUUGAAUUGCGCCAUUGCUGCACUUGGCACAGCGCAAAAGUUUGAUGGAAUUUAAAA_________________ ..(((((..................(((((.....)))))...(((((...((((......))))..))))).......))))).......................... ( -7.75 = -8.06 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:29 2011