| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,122,062 – 15,122,188 |
| Length | 126 |
| Max. P | 0.779714 |

| Location | 15,122,062 – 15,122,157 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 68.49 |
| Shannon entropy | 0.59895 |
| G+C content | 0.44784 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.58 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
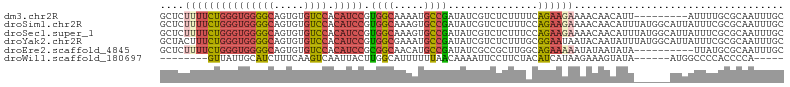
>dm3.chr2R 15122062 95 + 21146708 GCUCUUUUCUGGGUGGGGCAGUGUGUCCACAUCCGUGGCAAAAUGCCGAUAUCGUCUCUUUUCAGAAGAAAACAACAUU---------AUUUUGCGCAAUUUGC ((........(((((((((.....)))).)))))...((((((((.((....))....(((((....)))))......)---------)))))))))....... ( -23.50, z-score = -0.51, R) >droSim1.chr2R 13831345 104 + 19596830 GCUCUUUUCUGGGUGGGGCAGUGUGUCCACAUCCGUGGCAAAGUGCCGAUAUCGUCUCUUUCCAGAAGAAAACAACAUUUAUGGCAUUAUUUCGCGCAAUUUGC ((..(((((((((.((((((.(.((.((((....)))))).).))))(((...))).)).)))))))))..............((........))))....... ( -28.30, z-score = -0.90, R) >droSec1.super_1 12636669 104 + 14215200 GCUCUUUUCUGGGUGGGGCAGUGUGUCCACAUCCGUGGCAAAGUGCCGAUAUCGUCUCUUUCCAGAAGAAAACAACAUUUAUGGCAUUAUUUCGCGCAAUUUGC ((..(((((((((.((((((.(.((.((((....)))))).).))))(((...))).)).)))))))))..............((........))))....... ( -28.30, z-score = -0.90, R) >droYak2.chr2R 7082665 104 - 21139217 GCUACUUUCUGGGUGGGGCAGUGUGUCCACAUCCGUGGCGAAAUGCCGAUAUCGUCUCUUUGCGGAAUAAUACAAUAUUUAUGGCAUUAUUUCGCGCAAUUUGC (((((.....(((((((((.....)))).))))))))))(((.(((((....)).......((((((((((.((.......))..))))))))))))).))).. ( -31.20, z-score = -1.58, R) >droEre2.scaffold_4845 9321558 94 + 22589142 GCUCUUUUCUGGGUGGGGCAGUGUGUCCACAUCCGCGGCAACAUGCCGAUAUCGCCGCUUGGCAGAAAAAUAUAAUAUA----------UUAUGCGCAAUUUGC ((..(((((((((((((((.....)))).))))).((((.....)))).....(((....))))))))).(((((....----------))))).))....... ( -28.20, z-score = -0.55, R) >droWil1.scaffold_180697 1162317 85 - 4168966 --------GUUAUUGCAUCUUUCAAGUCAAUUACUUGGCAUUUUUUAACAAAAUUCCUUCUACAUCAUAAGAAAGUAUA------AUGGCCCCACCCCA----- --------(((((((...(((((((((.....))))((.(((((.....))))).)).............)))))..))------))))).........----- ( -10.90, z-score = -1.18, R) >consensus GCUCUUUUCUGGGUGGGGCAGUGUGUCCACAUCCGUGGCAAAAUGCCGAUAUCGUCUCUUUCCAGAAGAAAACAACAUU______AUUAUUUCGCGCAAUUUGC ....(((((((((((((((.....)))).))))).((((.....))))...............))))))................................... (-13.55 = -14.58 + 1.03)

| Location | 15,122,093 – 15,122,188 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.39 |
| Shannon entropy | 0.61136 |
| G+C content | 0.43723 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -11.50 |
| Energy contribution | -14.03 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
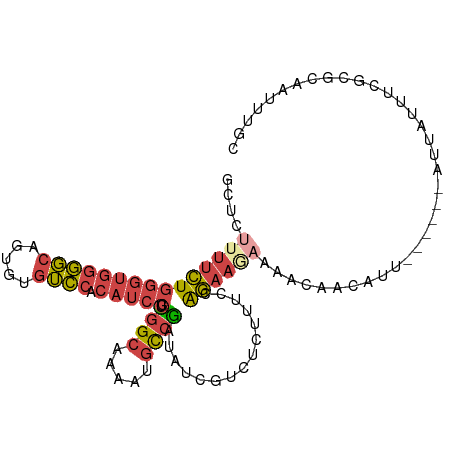
>dm3.chr2R 15122093 95 - 21146708 AUUCCGU---UUGCGGUCGGAAUUUAGCGGGACUGCAAAUUGCGCAAAAU---------AAUGUUGUUUUCUUCUGAAAAGAGACGAUAUCGGCAUUUUGCCACGGA- ..(((((---((((((((............))))))))).((.(((((((---------..((.((((.(((.((....))))).)))).))..)))))))))))))- ( -29.40, z-score = -1.98, R) >droSim1.chr2R 13831376 104 - 19596830 AUUCCGU---UUGCGGUCGGCAUUUAGCGGGACUGCAAAUUGCGCGAAAUAAUGCCAUAAAUGUUGUUUUCUUCUGGAAAGAGACGAUAUCGGCACUUUGCCACGGA- ..(((((---((((((((.((.....))..))))))))).((.(((((....((((....(((((((((..((....))..))))))))).)))).)))))))))))- ( -33.00, z-score = -1.73, R) >droSec1.super_1 12636700 104 - 14215200 AUUCCGU---UUGCGGUCGGCAUUUAGCGGGACUGCAAAUUGCGCGAAAUAAUGCCAUAAAUGUUGUUUUCUUCUGGAAAGAGACGAUAUCGGCACUUUGCCACGGA- ..(((((---((((((((.((.....))..))))))))).((.(((((....((((....(((((((((..((....))..))))))))).)))).)))))))))))- ( -33.00, z-score = -1.73, R) >droYak2.chr2R 7082696 107 + 21139217 AUUCCGUCGCUUGCGGUCGGAAUUUAGCGGGACUGCAAAUUGCGCGAAAUAAUGCCAUAAAUAUUGUAUUAUUCCGCAAAGAGACGAUAUCGGCAUUUCGCCACGGA- ..(((((.((((((((((............))))))))..((((((.((((((((.((....)).)))))))).))).......((....)))))....)).)))))- ( -33.00, z-score = -1.28, R) >droEre2.scaffold_4845 9321589 94 - 22589142 AUUCCGU---UUGCGGUGGGAAUUCAGCGGGACGGCAAAUUGCGCAUAAU----------AUAUUAUAUUUUUCUGCCAAGCGGCGAUAUCGGCAUGUUGCCGCGGA- ..(((..---....((..((((....(((.((.......)).)))((((.----------...))))...))))..))..((((((((((....)))))))))))))- ( -29.70, z-score = -0.43, R) >droWil1.scaffold_180697 1162349 84 + 4168966 --------------------AUUUUUUUUGCAAAGCAAAUGACGCUAUGUUUGG--GGUGGGGCCAUUAUACUUUCUUAUGAUGUAGAA--GGAAUUUUGUUAAAAAA --------------------..((((((.(((((((...(.((.(((....)))--.)).).)).......((((((........))))--))...))))).)))))) ( -15.80, z-score = -0.55, R) >consensus AUUCCGU___UUGCGGUCGGAAUUUAGCGGGACUGCAAAUUGCGCGAAAUAAUG__AUAAAUGUUGUUUUCUUCUGCAAAGAGACGAUAUCGGCAUUUUGCCACGGA_ ..(((((...((((((((............))))))))......................(((((((.(((((.....)))))))))))).(((.....)))))))). (-11.50 = -14.03 + 2.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:23 2011