| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,100,710 – 15,100,832 |
| Length | 122 |
| Max. P | 0.584556 |

| Location | 15,100,710 – 15,100,832 |
|---|---|
| Length | 122 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.59678 |
| G+C content | 0.37143 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -13.35 |
| Energy contribution | -15.05 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
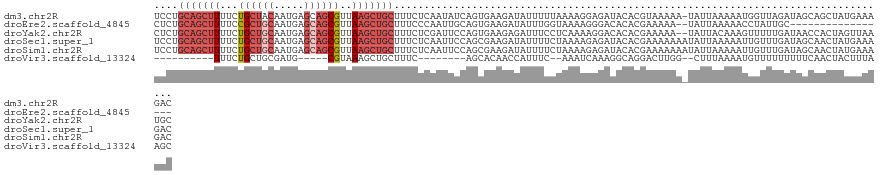
>dm3.chr2R 15100710 122 + 21146708 UCCUGCAGCUUUUCUGCUACAAUGAGCAGCGUUAAGCUGCUUUCUCAAUAUCAGUGAAGAUAUUUUUAAAAGGAGAUACACGUAAAAA-UAUUAAAAAUGGUUAGAUAGCAGCUAUGAAAGAC ........((((((((((......))))).....(((((((.(((....((((.....((((((((((...(........).))))))-)))).....)))).))).)))))))..))))).. ( -29.30, z-score = -1.55, R) >droEre2.scaffold_4845 9300435 104 + 22589142 CUCUGCAGCUUUUCCGCUGCAAUGAGCAGCGUUAAGCUGCUUUCCCAAUUGCAGUGAAGAUAUUUGGUAAAAGGGACACACGAAAAA--UAUUAAAAACCUAUUGC----------------- ....(((((((...((((((.....))))))..)))))))..((((..((((...(((....))).))))..))))...........--.................----------------- ( -29.20, z-score = -2.18, R) >droYak2.chr2R 7060765 121 - 21139217 CUCUGCAGCUUUUCUGCUGCAAUGAGCAGCGUUAAGCUGCUUUCUCGAUUCCAGUGAAGAGAUUUCCUCAAAAGGACACACGAAAAA--UAUUACAAAGUUUUUGAUAACCACUAGUUAAUGC ...((((((......))))))..(((((((.....)))))))..........((((........((((....))))....((((((.--..........)))))).....))))......... ( -27.90, z-score = -0.80, R) >droSec1.super_1 12615438 123 + 14215200 UCCUGCAGCUUUUCUGCUGCAAUGAGCAGCGUUAAGCUGCUUUCUCAAUUCCAGCGAAGAUAUUUUCUAAAAGAGAUACACGAAAAAAAUAUUAAAAAUUGUUUGAUAGCAACUAUGAAAGAC ...((((((......))))))..(((((((.....)))))))...........((........((((.....)))).............(((((((.....)))))))))............. ( -26.00, z-score = -0.71, R) >droSim1.chr2R 13810859 123 + 19596830 UCCUGCAGCUUUUCUGCUGCAAUGAGCAGCGUUAAGCUGCUUUCUCAAUUCCAGCGAAGAUAUUUUCUAAAAGAGAUACACGAAAAAAAUAUUAAAAAUUGUUUGAUAGCAACUAUGAAAGAC ...((((((......))))))..(((((((.....)))))))...........((........((((.....)))).............(((((((.....)))))))))............. ( -26.00, z-score = -0.71, R) >droVir3.scaffold_13324 2738609 96 + 2960039 ----------UUUCUGCUGCGAUG-----CGUAAAGCUGCUUUC--------AGCACAACCAUUUC--AAAUCAAAGGCAGGACUUGG--CUUUAAAAUGUUUUUUUUUCAACUACUUUAAGC ----------.(((((((..((((-----(.....))(((....--------.)))..........--..)))...)))))))....(--(((......(((........)))......)))) ( -15.10, z-score = 1.21, R) >consensus UCCUGCAGCUUUUCUGCUGCAAUGAGCAGCGUUAAGCUGCUUUCUCAAUUCCAGCGAAGAUAUUUUCUAAAAGAGAUACACGAAAAAA_UAUUAAAAAUUGUUUGAUAGCAACUAUGAAAGAC ....(((((((...((((((.....))))))..)))))))................................................................................... (-13.35 = -15.05 + 1.70)
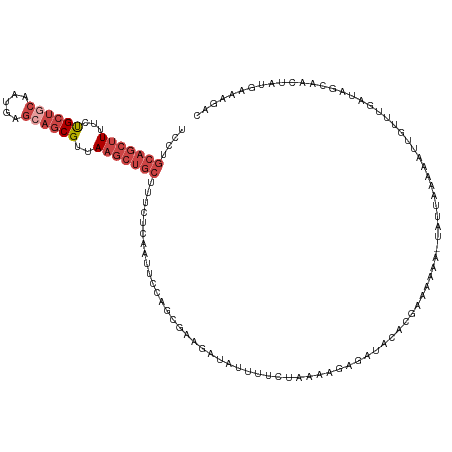

| Location | 15,100,710 – 15,100,832 |
|---|---|
| Length | 122 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.59678 |
| G+C content | 0.37143 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.67 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
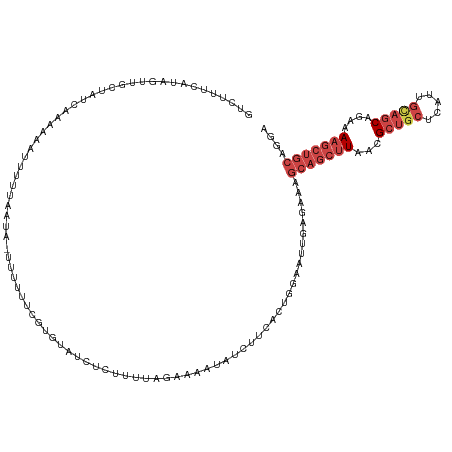
>dm3.chr2R 15100710 122 - 21146708 GUCUUUCAUAGCUGCUAUCUAACCAUUUUUAAUA-UUUUUACGUGUAUCUCCUUUUAAAAAUAUCUUCACUGAUAUUGAGAAAGCAGCUUAACGCUGCUCAUUGUAGCAGAAAAGCUGCAGGA (..((((..(((((((.(((...........(((-(......)))).............((((((......)))))).))).)))))))....(((((.....))))).))))..)....... ( -28.00, z-score = -1.81, R) >droEre2.scaffold_4845 9300435 104 - 22589142 -----------------GCAAUAGGUUUUUAAUA--UUUUUCGUGUGUCCCUUUUACCAAAUAUCUUCACUGCAAUUGGGAAAGCAGCUUAACGCUGCUCAUUGCAGCGGAAAAGCUGCAGAG -----------------.................--....((.....((((..........................))))..(((((((..((((((.....))))))...))))))).)). ( -27.97, z-score = -1.47, R) >droYak2.chr2R 7060765 121 + 21139217 GCAUUAACUAGUGGUUAUCAAAAACUUUGUAAUA--UUUUUCGUGUGUCCUUUUGAGGAAAUCUCUUCACUGGAAUCGAGAAAGCAGCUUAACGCUGCUCAUUGCAGCAGAAAAGCUGCAGAG ..((((...(((...........)))...)))).--(((((((....(((...((((((....))))))..)))..)))))))(((((((...(((((.....)))))....))))))).... ( -36.30, z-score = -2.52, R) >droSec1.super_1 12615438 123 - 14215200 GUCUUUCAUAGUUGCUAUCAAACAAUUUUUAAUAUUUUUUUCGUGUAUCUCUUUUAGAAAAUAUCUUCGCUGGAAUUGAGAAAGCAGCUUAACGCUGCUCAUUGCAGCAGAAAAGCUGCAGGA (..((((..(((((((.((...(((((((..(((((((((....(.....)....))))))))).......))))))).)).)))))))....(((((.....))))).))))..)....... ( -27.50, z-score = -0.37, R) >droSim1.chr2R 13810859 123 - 19596830 GUCUUUCAUAGUUGCUAUCAAACAAUUUUUAAUAUUUUUUUCGUGUAUCUCUUUUAGAAAAUAUCUUCGCUGGAAUUGAGAAAGCAGCUUAACGCUGCUCAUUGCAGCAGAAAAGCUGCAGGA (..((((..(((((((.((...(((((((..(((((((((....(.....)....))))))))).......))))))).)).)))))))....(((((.....))))).))))..)....... ( -27.50, z-score = -0.37, R) >droVir3.scaffold_13324 2738609 96 - 2960039 GCUUAAAGUAGUUGAAAAAAAAACAUUUUAAAG--CCAAGUCCUGCCUUUGAUUU--GAAAUGGUUGUGCU--------GAAAGCAGCUUUACG-----CAUCGCAGCAGAAA---------- (((((((((.(((........)))))))).)))--)((((((........)))))--)......((.((((--------(...((........)-----)....))))).)).---------- ( -18.90, z-score = 0.49, R) >consensus GUCUUUCAUAGUUGCUAUCAAAAAAUUUUUAAUA_UUUUUUCGUGUAUCUCUUUUAGAAAAUAUCUUCACUGGAAUUGAGAAAGCAGCUUAACGCUGCUCAUUGCAGCAGAAAAGCUGCAGGA ...................................................................................(((((((...(((((.....)))))....))))))).... (-13.11 = -14.67 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:17 2011