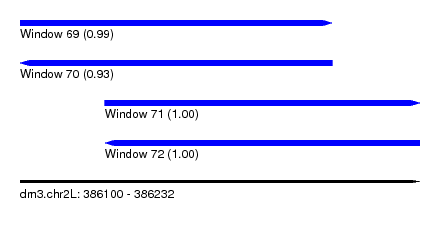
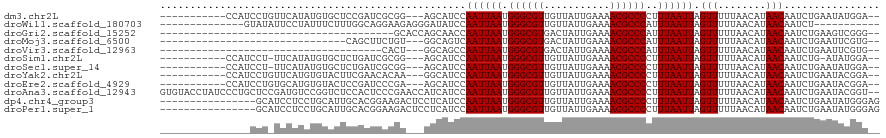
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 386,100 – 386,232 |
| Length | 132 |
| Max. P | 0.999196 |

| Location | 386,100 – 386,203 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.46391 |
| G+C content | 0.38943 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
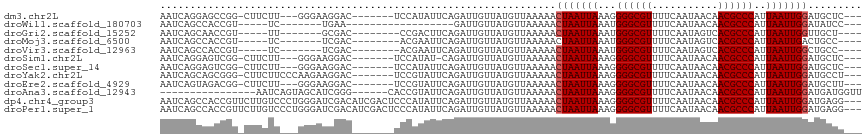
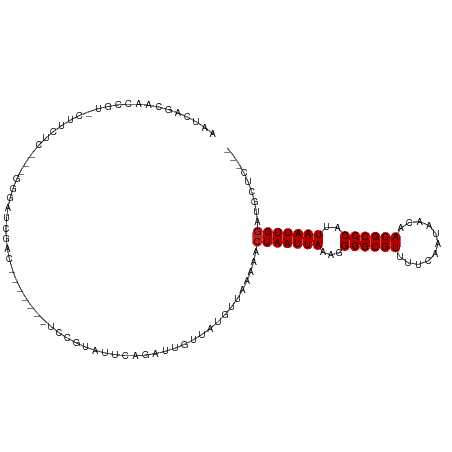
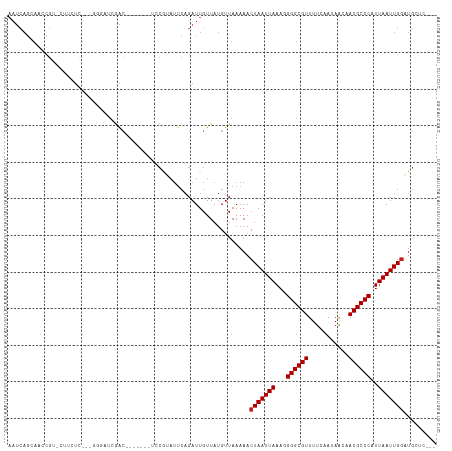
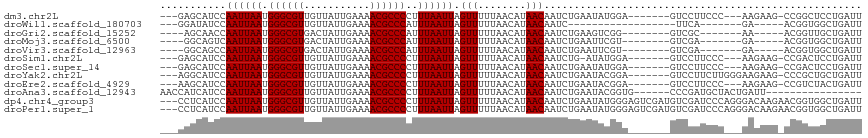
>dm3.chr2L 386100 103 + 23011544 AAUCAGGAGCCGG-CUUCUU---GGGAAGGAC-------UCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCUC--- .....((((((..-((....---.))..)).)-------)))........................((((((((..(((((((.........))))))).))))))))......--- ( -26.80, z-score = -1.74, R) >droWil1.scaffold_180703 2556248 84 - 3946847 AAUCAGCCACCGU-----UC-------UGAA------------------GAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUAUCC--- ..((((.......-----.)-------))).------------------.................((((((((.((((((((.........))))))))))))))))......--- ( -20.50, z-score = -3.10, R) >droGri2.scaffold_15252 6699138 93 + 17193109 AAUCAGCAACCGU-----UU-------GCGAC--------CCGACUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGGUUGCU---- ....(((((((((-----((-------..(((--------..(((........)))..)))..)))).((((((.(((((((...(....)..))))))))))))))))))))---- ( -29.10, z-score = -4.27, R) >droMoj3.scaffold_6500 7965954 93 - 32352404 AAUCAGCCACCGU-----UC-------UCGAC--------ACGAAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGACUGCC---- ...(((.....((-----(.-------(..((--------(..(((.......))).)))..).)))(((((((.(((((((...(....)..)))))))))))))).)))..---- ( -16.90, z-score = -0.94, R) >droVir3.scaffold_12963 18777542 93 + 20206255 AAUCAGCCACCGU-----UC-------UCGAC--------ACGAAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGGCUGCC---- ...((((((..((-----(.-------(..((--------(..(((.......))).)))..).)))......(((((((((...(....)..)))))))))...))))))..---- ( -23.70, z-score = -2.84, R) >droSim1.chr2L 392519 102 + 22036055 AAUCAGGAGUCGG-CUUCUU---GGGAAGGAC-------UCCAUAU-CAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCUC--- .....((((((..-((((..---..)))))))-------)))....-...................((((((((..(((((((.........))))))).))))))))......--- ( -29.70, z-score = -3.04, R) >droSec1.super_14 369572 103 + 2068291 AAUCAGGAGUCGG-CUUCUU---GGGAAGGAC-------UCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCUC--- .....((((((..-((((..---..)))))))-------)))........................((((((((..(((((((.........))))))).))))))))......--- ( -29.70, z-score = -3.08, R) >droYak2.chr2L 369549 106 + 22324452 AAUCAGCAGCGGG-CUUCUUCCCAAGAAGGAC-------UCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCCU--- .....(((((((.-.((((((....)))))).-------.))))......................((((((((..(((((((.........))))))).)))))))).)))..--- ( -31.30, z-score = -2.95, R) >droEre2.scaffold_4929 434002 103 + 26641161 AAUCAGUAGACGG-CUUCUU---GGGAAGGAC-------UCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCUU--- ((((.(...((((-..((((---....)))).-------.))))...).)))).............((((((((..(((((((.........))))))).))))))))......--- ( -26.40, z-score = -2.01, R) >droAna3.scaffold_12943 167297 95 - 5039921 ----------------AAUCAGUAGCAUCGGG------CACCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAUGGUU ----------------.((((((((((((.((------........)).)).))))))........((((((((..(((((((.........))))))).)))))))).)))).... ( -22.50, z-score = -1.64, R) >dp4.chr4_group3 10439141 114 - 11692001 AAUCAGCCACCGUUCUUGUCCCUGGGAUCGACAUCGACUCCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAGG--- ..............((..(((.((((((((....))).))))).........................((((((..(((((((.........))))))).)))))))))..)).--- ( -30.10, z-score = -2.00, R) >droPer1.super_1 7559016 114 - 10282868 AAUCAGCCACCGUUCUUGUCCCUGGGAUCGACAUCGACUCCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAGG--- ..............((..(((.((((((((....))).))))).........................((((((..(((((((.........))))))).)))))))))..)).--- ( -30.10, z-score = -2.00, R) >consensus AAUCAGCAACCGU_CUUCUC___GGGAUCGAC_______UCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCUC___ ..................................................................(((((((...((((((...........))))))..)))))))......... (-14.35 = -14.43 + 0.08)
| Location | 386,100 – 386,203 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.46391 |
| G+C content | 0.38943 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 386100 103 - 23011544 ---GAGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGA-------GUCCUUCCC---AAGAAG-CCGGCUCCUGAUU ---........((((((.(((((((.........)))))))..)))))).(((........)))...........(((-------(((((((..---..))))-..))))))..... ( -25.20, z-score = -2.13, R) >droWil1.scaffold_180703 2556248 84 + 3946847 ---GGAUAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUC------------------UUCA-------GA-----ACGGUGGCUGAUU ---((....))((((((((((((((.........)))))))).)))))).(((........)))....------------------.(((-------(.-----.......)))).. ( -19.80, z-score = -2.07, R) >droGri2.scaffold_15252 6699138 93 - 17193109 ----AGCAACCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAGUCGG--------GUCGC-------AA-----ACGGUUGCUGAUU ----((((((((((((((((((((...........))))))).))))))................(((((....)))--------))...-------..-----..))))))).... ( -28.80, z-score = -3.69, R) >droMoj3.scaffold_6500 7965954 93 + 32352404 ----GGCAGUCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAUUCGU--------GUCGA-------GA-----ACGGUGGCUGAUU ----..((((((((((((((((((...........))))))).)))))).(((((..((((..............))--------))..)-------))-----))...)))))... ( -27.54, z-score = -3.17, R) >droVir3.scaffold_12963 18777542 93 - 20206255 ----GGCAGCCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAUUCGU--------GUCGA-------GA-----ACGGUGGCUGAUU ----..((((((((((((((((((...........))))))).)))))).(((((..((((..............))--------))..)-------))-----))...)))))... ( -30.24, z-score = -3.90, R) >droSim1.chr2L 392519 102 - 22036055 ---GAGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUG-AUAUGGA-------GUCCUUCCC---AAGAAG-CCGACUCCUGAUU ---....(((.((((((.(((((((.........)))))))..)))))).(((........))).....)-))..(((-------(((((((..---..))))-..))))))..... ( -26.00, z-score = -3.13, R) >droSec1.super_14 369572 103 - 2068291 ---GAGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGA-------GUCCUUCCC---AAGAAG-CCGACUCCUGAUU ---........((((((.(((((((.........)))))))..)))))).(((........)))...........(((-------(((((((..---..))))-..))))))..... ( -25.80, z-score = -3.14, R) >droYak2.chr2L 369549 106 - 22324452 ---AGGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA-------GUCCUUCUUGGGAAGAAG-CCCGCUGCUGAUU ---.(((.(((((((((.(((((((.........)))))))..)))))).(((........)))...........)))-------.((((....))))....)-))........... ( -26.40, z-score = -1.17, R) >droEre2.scaffold_4929 434002 103 - 26641161 ---AAGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA-------GUCCUUCCC---AAGAAG-CCGUCUACUGAUU ---........((((((.(((((((.........)))))))..)))))).(((........)))((((.....((((.-------.((......---..))..-)))).....)))) ( -22.10, z-score = -1.99, R) >droAna3.scaffold_12943 167297 95 + 5039921 AACCAUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGUG------CCCGAUGCUACUGAUU---------------- .....(((...((((((.(((((((.........)))))))..)))))).(((........)))....)))...(((((------(.....)).))))...---------------- ( -19.10, z-score = -1.55, R) >dp4.chr4_group3 10439141 114 + 11692001 ---CCUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGGAGUCGAUGUCGAUCCCAGGGACAAGAACGGUGGCUGAUU ---..(((.((((((((.(((((((.........)))))))..)))))).............((..(((.....(((((.(((....))))))))......)))...)))).))).. ( -30.70, z-score = -1.35, R) >droPer1.super_1 7559016 114 + 10282868 ---CCUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGGAGUCGAUGUCGAUCCCAGGGACAAGAACGGUGGCUGAUU ---..(((.((((((((.(((((((.........)))))))..)))))).............((..(((.....(((((.(((....))))))))......)))...)))).))).. ( -30.70, z-score = -1.35, R) >consensus ___GAGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA_______GUCCAUCCC___AAGAAG_ACGGUUGCUGAUU ...........((((((.((((((...........))))))..)))))).(((........)))..................................................... (-11.38 = -11.38 + 0.00)
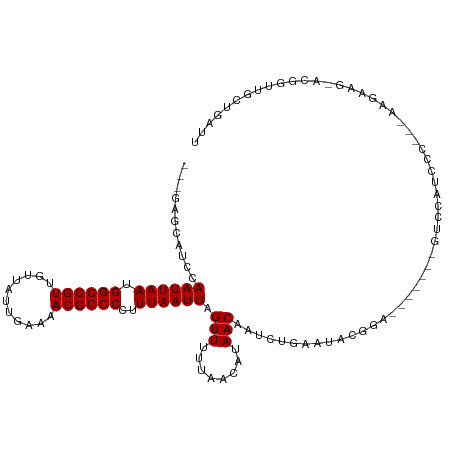
| Location | 386,128 – 386,232 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.60206 |
| G+C content | 0.38613 |
| Mean single sequence MFE | -26.60 |
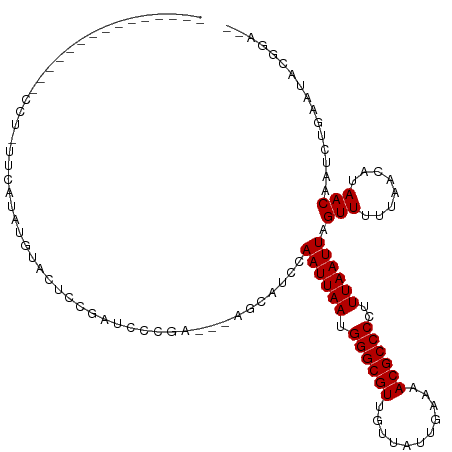
| Consensus MFE | -14.35 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 386128 104 + 23011544 --UCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCU---CCGCGAUCGGAGCACAUAUGAACAGGAUGG----------- --........((..(((((((((......((((((((..(((((((.........))))))).)))))))).((((---(((....))))))).))))).))))..)).----------- ( -32.90, z-score = -4.16, R) >droWil1.scaffold_180703 2556266 95 - 3946847 -----------AGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUAUCCCUCUUCCUGCCAAAGAAAUAGGAAUAUAC-------------- -----------.......((((((.....((((((((.((((((((.........)))))))))))))))).....(((.((.((.....)).)).))))))))).-------------- ( -23.10, z-score = -3.25, R) >droGri2.scaffold_15252 6699157 79 + 17193109 --CCCGACUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGGUUGCUGGUGC--------------------------------------- --.(((((........)))........((((((((((.(((((((...(....)..)))))))))))))))))...))...--------------------------------------- ( -21.10, z-score = -2.33, R) >droMoj3.scaffold_6500 7965973 84 - 32352404 --CACGAAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGACUGCC---ACAGAAGCUG------------------------------- --........(((.................(((((((.(((((((...(....)..)))))))))))))).(((..---.)))...)))------------------------------- ( -15.20, z-score = -0.86, R) >droVir3.scaffold_12963 18777561 78 + 20206255 --CACGAAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAUGGGCGUUUUCAAUAGUCACGCCCAUUAAUUGGCUGCC---AGUG------------------------------------- --(((.....(((.(((......)))...((((((((.(((((((...(....)..))))))))))))))))))..---.)))------------------------------------- ( -17.90, z-score = -1.83, R) >droSim1.chr2L 392547 102 + 22036055 --UCCAUAU-CAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCU---CCGCGAUCAGAGCACAUAUGAA-AGGAUGG----------- --.((((.(-((...(((...........((((((((..(((((((.........))))))).))))))))..(((---(........)))))))..))).-...))))----------- ( -26.80, z-score = -2.62, R) >droSec1.super_14 369600 103 + 2068291 --UCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCU---CCGCGAUCAGAGCACAUAUGAA-AGGAUGG----------- --.((((.((((...(((...........((((((((..(((((((.........))))))).))))))))..(((---(........)))))))..))))-...))))----------- ( -28.30, z-score = -3.05, R) >droYak2.chr2L 369580 104 + 22324452 --UCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCC---UUGUGUUCGAAGUACACAUGAACAGGAUGG----------- --(((...((((.................((((((((..(((((((.........))))))).)))))))).....---.(((((.......)))))))))..)))...----------- ( -25.70, z-score = -1.74, R) >droEre2.scaffold_4929 434030 104 + 26641161 --UCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCU---UCGGGAUCGGAGUACACAUGCACAGGAUGG----------- --........((..(((((((((......((((((((..(((((((.........))))))).)))))))).((((---(((....))))))).))))).))))..)).----------- ( -30.60, z-score = -3.22, R) >droAna3.scaffold_12943 167314 118 - 5039921 --ACCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAUGGUUCGGGAGUGGAGACCGGACAUCGGAGCAGGGAUAGGUACAC --(((.(((((....((((..........((((((((..(((((((.........))))))).)))))))).(((((.(((((.........)))))))))).)))).)))))))).... ( -34.60, z-score = -2.81, R) >dp4.chr4_group3 10439178 104 - 11692001 CUCCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAGGAGUCUUCCGUGCAAUGCAGGAGGAUGC---------------- (((((((.....(((......))).....((((((((..(((((((.........))))))).))))))))))).))))((((((.(((...)))))))))...---------------- ( -31.50, z-score = -3.00, R) >droPer1.super_1 7559053 104 - 10282868 CUCCCAUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGAGGAGUCUUCCGUGCAAUGCAGGAGGAUGC---------------- (((((((.....(((......))).....((((((((..(((((((.........))))))).))))))))))).))))((((((.(((...)))))))))...---------------- ( -31.50, z-score = -3.00, R) >consensus __CCCGUAUUCAGAUUGUUAUGUUAAAAACUAAUUAAAGGGGCGUUUUCAAUAACAACGCCCAUUAAUUGGAUGCU___UCGCGAGCGGAGUACAUAUGAA_AGG_______________ .............................(((((((...((((((...........))))))..)))))))................................................. (-14.35 = -14.43 + 0.08)


| Location | 386,128 – 386,232 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.60206 |
| G+C content | 0.38613 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 386128 104 - 23011544 -----------CCAUCCUGUUCAUAUGUGCUCCGAUCGCGG---AGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGA-- -----------...((((((((((((((((((((....)))---)))....((((((.(((((((.........)))))))..))))))........)))))......)))))).)))-- ( -33.80, z-score = -4.55, R) >droWil1.scaffold_180703 2556266 95 + 3946847 --------------GUAUAUUCCUAUUUCUUUGGCAGGAAGAGGGAUAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCU----------- --------------(.((((((((.(((((......))))))))))))).)((((((((((((((.........)))))))).)))))).(((........))).....----------- ( -26.70, z-score = -3.57, R) >droGri2.scaffold_15252 6699157 79 - 17193109 ---------------------------------------GCACCAGCAACCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAGUCGGG-- ---------------------------------------...((.((....(((((((((((((...........))))))).)))))).(((........)))........)).)).-- ( -18.40, z-score = -2.31, R) >droMoj3.scaffold_6500 7965973 84 + 32352404 -------------------------------CAGCUUCUGU---GGCAGUCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAUUCGUG-- -------------------------------..(((.....---)))....(((((((((((((...........))))))).)))))).(((........)))..............-- ( -17.20, z-score = -1.13, R) >droVir3.scaffold_12963 18777561 78 - 20206255 -------------------------------------CACU---GGCAGCCAAUUAAUGGGCGUGACUAUUGAAAACGCCCAUUUAAUUAGUUUUUAACAUAACAAUCUGAAUUCGUG-- -------------------------------------(((.---..(((..(((((((((((((...........))))))).)))))).(((........)))...))).....)))-- ( -16.80, z-score = -1.81, R) >droSim1.chr2L 392547 102 - 22036055 -----------CCAUCCU-UUCAUAUGUGCUCUGAUCGCGG---AGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUG-AUAUGGA-- -----------.......-(((((((((((((((....)))---)))))..((((((.(((((((.........)))))))..)))))).....................-)))))))-- ( -28.50, z-score = -3.20, R) >droSec1.super_14 369600 103 - 2068291 -----------CCAUCCU-UUCAUAUGUGCUCUGAUCGCGG---AGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGA-- -----------((((...-(((((((((((((((....)))---)))....((((((.(((((((.........)))))))..))))))........)))))......)))).)))).-- ( -29.60, z-score = -3.64, R) >droYak2.chr2L 369580 104 - 22324452 -----------CCAUCCUGUUCAUGUGUACUUCGAACACAA---GGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA-- -----------...((((((((((((((.......))))).---.......((((((.(((((((.........)))))))..))))))...................)))))).)))-- ( -25.00, z-score = -2.34, R) >droEre2.scaffold_4929 434030 104 - 26641161 -----------CCAUCCUGUGCAUGUGUACUCCGAUCCCGA---AGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA-- -----------..(((..((((....))))...))).(((.---.......((((((.(((((((.........)))))))..)))))).(((........)))..........))).-- ( -22.00, z-score = -1.36, R) >droAna3.scaffold_12943 167314 118 + 5039921 GUGUACCUAUCCCUGCUCCGAUGUCCGGUCUCCACUCCCGAACCAUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGU-- ((((...............((((..(((.........)))...))))....((((((.(((((((.........)))))))..))))))......................))))...-- ( -21.80, z-score = -0.82, R) >dp4.chr4_group3 10439178 104 + 11692001 ----------------GCAUCCUCCUGCAUUGCACGGAAGACUCCUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGGAG ----------------...((.((((((...))).))).))((((.(((..((((((.(((((((.........)))))))..)))))).(((........))).........))))))) ( -25.80, z-score = -2.27, R) >droPer1.super_1 7559053 104 + 10282868 ----------------GCAUCCUCCUGCAUUGCACGGAAGACUCCUCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUAUGGGAG ----------------...((.((((((...))).))).))((((.(((..((((((.(((((((.........)))))))..)))))).(((........))).........))))))) ( -25.80, z-score = -2.27, R) >consensus _______________CCU_UUCAUAUGUACUCCGAUCCCGA___AGCAUCCAAUUAAUGGGCGUUGUUAUUGAAAACGCCCCUUUAAUUAGUUUUUAACAUAACAAUCUGAAUACGGA__ ...................................................((((((.((((((...........))))))..)))))).(((........)))................ (-11.38 = -11.38 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:39 2011