
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,054,462 – 15,054,571 |
| Length | 109 |
| Max. P | 0.776917 |

| Location | 15,054,462 – 15,054,571 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.19 |
| Shannon entropy | 0.57077 |
| G+C content | 0.51419 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -12.42 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15054462 109 + 21146708 ----UUGACCAAACGUGGUCAAAACAAUUGGCCCCGCAGUUCAAAUACCUUUUGGCCAGCCGAAGGCGGG------UGGCGAGUAAACUGACCACCGACUAUAUUUGCAGGCCAAUCAU ----(((((((....)))))))....(((((((..((((........((((((((....))))))).)((------(((..((....))..)))))........)))).)))))))... ( -39.60, z-score = -3.19, R) >droPer1.super_4 5205836 110 + 7162766 CCGCGGGUCCUGGACUGGUCAAAACACUUGGUGGCGGGGUUAAAGUGCCUCUCACAUAGGCCGAGCAAAUUA----CUCCGCACAAA---AGCGUGGCAUUCAUUUGCCGACCAAUC-- (((.(....))))..(((((...((.(((..(((((((((....((((((.......))))...)).....)----)))))).)).)---)).))((((......)))))))))...-- ( -34.90, z-score = -0.67, R) >dp4.chr3 9887823 110 + 19779522 CCGCGGGUCCUGGACUGGUCAAAACACUUGGUGGCGGGGUUAAAGUGCCUCUCACAUAGGCCGAGCAAAUUG----CUCCGCACAAA---ACCGUGGCAUUCAUUUGCCGACCAAUC-- (((.(....))))..(((((.......(((((((.(((((......)))))))))....((.((((.....)----))).)).))).---.....((((......)))))))))...-- ( -35.40, z-score = -0.48, R) >droEre2.scaffold_4845 9254399 104 + 22589142 ----GUGCCCAAACGUGGUCAAAACAAUUGGCCCCGAAGUUCAAAUACCUUG-----AGGAAAAGGCUGG------UGGCGAGUAAAUUGACCGCCAGCUAUAUUUGCAGGCCCAUCAU ----(.(((....((.((((((.....)))))).)).....((((((((...-----.))....((((((------((((((.....))).))))))))).))))))..))).)..... ( -34.80, z-score = -2.36, R) >droYak2.chr2R 7014042 115 - 21139217 ----CUGCCCAAACGUGGUCAAAACAAUUGGACCCGAAGUUCAAAUACCUUUUGGCCAAGCAGAGGUGAGCGCGGUUGGCGAGUAAACUGACCCCCGACUAUAUUUGCAGGCCAAUCAU ----..(((......(((((((((...((((((.....)))))).....))))))))).(((((.(((..((.(((..(........)..)))..))..))).))))).)))....... ( -31.10, z-score = -0.40, R) >droSim1.chr2R 13758769 115 + 19596830 ----GUGACCAAACGUGGUCAAAACAAUUGGCCCCGAAGUUCAAAUACCUUUUGGCCAGCCGAAGGUAAAUGCGGUUGGCGAGUCAACUGGCCGCCGACUAUAUUUGCAGGCCAAUCAU ----.((((((....)))))).....(((((((................((((((....))))))(((((((..(((((((.(((....))))))))))..))))))).)))))))... ( -46.10, z-score = -4.18, R) >droSec1.super_1 12567263 115 + 14215200 ----GUGACCAAACGUGGUCAAAACAAUUGGCCCCGAAGUUCAAAUACCUUUUGGCCAGCCGAAGGUAAAUGCGGUUGGCGAGUAAACUGGCCGCCGACUAUAUUUGCAGGCCAAUCAU ----.((((((....)))))).....(((((((................((((((....))))))(((((((..(((((((.((......)))))))))..))))))).)))))))... ( -43.60, z-score = -3.56, R) >consensus ____GUGACCAAACGUGGUCAAAACAAUUGGCCCCGAAGUUCAAAUACCUUUUGGCCAGGCGAAGGUAAGUG____UGGCGAGUAAACUGACCGCCGACUAUAUUUGCAGGCCAAUCAU ...............(((((.......(((....)))....((((((((((...........))))..........(((((.((......)))))))....))))))..)))))..... (-12.42 = -12.14 + -0.28)

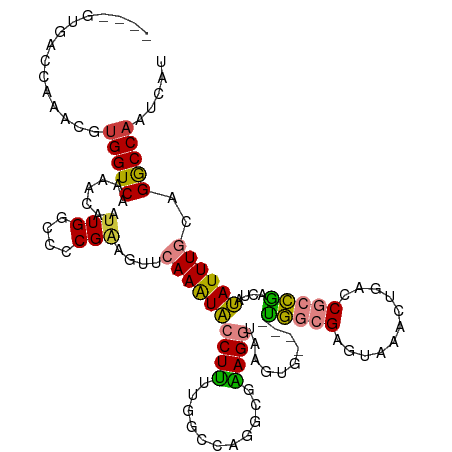
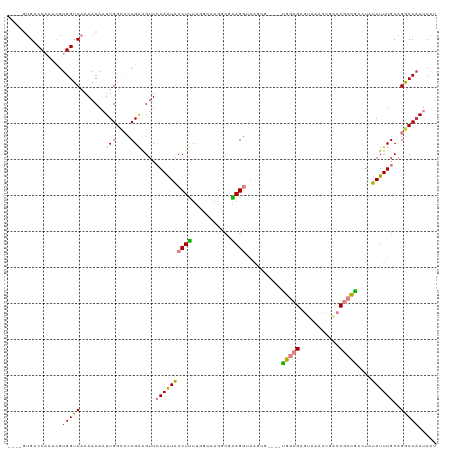
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:07 2011