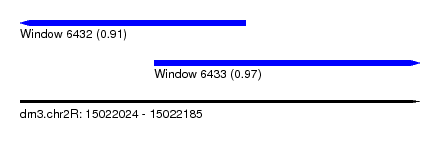
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 15,022,024 – 15,022,185 |
| Length | 161 |
| Max. P | 0.965760 |

| Location | 15,022,024 – 15,022,115 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.53013 |
| G+C content | 0.52058 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.89 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
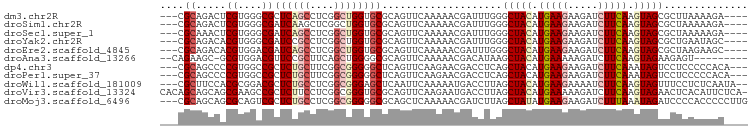
>dm3.chr2R 15022024 91 - 21146708 ---CGCAGACUCGUGGGCGCUCAGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCGCUUAAAAGA---- ---..((((.((((..(((((((((....))))).)))).........)))))))).(((((.(((((.....))))).)))))..........---- ( -33.20, z-score = -2.12, R) >droSim1.chr2R 13726100 91 - 19596830 ---CGCAGACUCGUGGGCGAUCAAGCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCGCUAAAAAGA---- ---.......((((((((......))))(((((.....))))).....))))((((((((((.(((((.....))))).))))).)))))....---- ( -28.40, z-score = -1.38, R) >droSec1.super_1 12535084 91 - 14215200 ---CGCAAACUCGUGGGCGAUCAGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCGCUAAAAAGA---- ---.......((((..(((((((((....)))))).))).........))))((((((((((.(((((.....))))).))))).)))))....---- ( -32.20, z-score = -2.66, R) >droYak2.chr2R 6980912 91 + 21139217 ---CGCAGACACGUGGGCGAUCCGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCGCUGAAUAGC---- ---..(((...((.(((((...))))))).)))..((((.(((((((.....)))))))(((.(((((.....))))).)))))))........---- ( -29.80, z-score = -1.10, R) >droEre2.scaffold_4845 9221716 91 - 22589142 ---CGCAGACACGUGGACGAUCAGCCUCGGCUGGUGCGCAGUUCAAAAACGAUUUGGGCUACAUGAAGAAGAUCUUCAAGUAGCGCUAAGAAGC---- ---.((.......(((((.((((((....)))))).....))))).......((((((((((.(((((.....))))).))))).)))))..))---- ( -29.40, z-score = -1.71, R) >droAna3.scaffold_13266 7128006 86 + 19884421 --CAGAAGC-GCGUGGACGUUCCGCUUCAGCUGGGGCGCAGUUCAAAAACGACAUAAGCUACAUGAAAAAGAUCUUCAAGUAGAAGAGU--------- --..(.(((-..((((.((((((((....)).))))))).(((....)))..)))..))).)..........(((((.....)))))..--------- ( -19.50, z-score = 0.01, R) >dp4.chr3 19745096 92 - 19779522 ---CGCAGCCCCGUGGCCGCUCUGCUUCGGCGGGGGCUCAGUUCAAGAACGACCUCAGCUACAUGAAGAAGAUCUUCAAAUAGUCCUCCCCCACA--- ---.((.(((....))).))...((....))(((((....(((....))).......((((..(((((.....)))))..))))...)))))...--- ( -29.70, z-score = -1.18, R) >droPer1.super_37 746411 92 - 760358 ---CGCAGCCCCGUGGCCGCUCUGCUUCGGCGGGGGCUCAGUUCAAGAACGACCUCAGCUACAUGAAGAAGAUCUUCAAAUAGUCCUCCCCCACA--- ---.((.(((....))).))...((....))(((((....(((....))).......((((..(((((.....)))))..))))...)))))...--- ( -29.70, z-score = -1.18, R) >droWil1.scaffold_181009 1702732 93 - 3585778 ---CGCUUCCACGCGGACGCUCUGCCUCGGCGGGAGCUCAAUUCAAAAAUGACCUUAGCUACAUGAAGAAAAUCUUCAAGUAGUUUCCUCUCAAUA-- ---.(((.((.(((.((.((...)).)).))))))))............(((....((((((.(((((.....))))).)))))).....)))...-- ( -27.70, z-score = -2.94, R) >droVir3.scaffold_13324 2669827 97 - 2960039 CACAGCAGCAGCGAAGCCGCUCUUCCUCGGCGGGUGCGCAGUUCAAGAAUGACCUUAGCUACAUGAAAAAGAUCUUCAAGUAGAACUCACAUUCUCA- ....(.(((.(((.(.(((((.......))))).).))).)))).((((((.......((((.((((.......)))).))))......))))))..- ( -24.42, z-score = -0.49, R) >droMoj3.scaffold_6496 12734529 95 + 26866924 ---CGCAGCAGCGCAGUCGCUCUGCCUCGGCGGGGGCGCAGCUCAAAAACGAUCUUAGCUAUAUGAAGAAGAUCUUUAAAUAGAUCCCCACCCCCUUG ---.((((.((((....))))))))...((.(((((...((.((......)).))...((((.(((((.....))))).)))).))))).))...... ( -27.30, z-score = -0.46, R) >consensus ___CGCAGCCACGUGGGCGCUCUGCCUCGGCUGGUGCGCAGUUCAAAAACGAUCUGAGCUACAUGAAGAAGAUCUUCAAGUAGCGCUCAAAAGC____ ....((.....((....))((((((....))))))))....................(((((.(((((.....))))).))))).............. (-16.39 = -16.89 + 0.50)

| Location | 15,022,078 – 15,022,185 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 68.63 |
| Shannon entropy | 0.66748 |
| G+C content | 0.64624 |
| Mean single sequence MFE | -48.99 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 15022078 107 + 21146708 UGCGCACCAGCCGAGGCUGAGCGCCCACGAGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGUUUGGGCGAGGGC------GAAUACCGCCCGUA .(((((((((((..((((...(.(((((.....(((((((((....))))))))).))))))...)).))..)))))))))....))..((((------(.....)))))... ( -52.40, z-score = -2.49, R) >droSim1.chr2R 13726154 107 + 19596830 UGCGCACCAGCCGAGCUUGAUCGCCCACGAGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUUAGGCGAGGGC------GAGUACCGCCCGUA ((((((((((((.(((((..((.(((((.....(((((((((....))))))))).))))))).))).)).).))))))))........((((------(.....)))))))) ( -53.10, z-score = -2.75, R) >droSec1.super_1 12535138 107 + 14215200 UGCGCACCAGCCGAGGCUGAUCGCCCACGAGUUUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUUGGGCGAGGGC------GAAAACCGCCCGUA ((((((((((((..((((..((.(((((....((((((((((....)))))))))))))))))..)).))..)))))))))..(((((.(...------.....))))))))) ( -56.40, z-score = -3.69, R) >droYak2.chr2R 6980966 107 - 21139217 UGCGCACCAGCCGAGGCGGAUCGCCCACGUGUCUGCGGCUCCAGUUGGAGCUGCGAGUGGGGAAAGGACUGAGGUUGGUGCUUUGGGGAGGGC------GAGUACCGCCCGUA ((((((((((((..(((...((.(((((.....(((((((((....))))))))).)))))))...).))..)))))))))........((((------(.....)))))))) ( -55.50, z-score = -2.96, R) >droEre2.scaffold_4845 9221770 107 + 22589142 UGCGCACCAGCCGAGGCUGAUCGUCCACGUGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUUGGGGGAGGGC------GAAUAACGCCCGUA ((((((((((((..((((..((.(((((.....(((((((((....))))))))).)))))))..)).))..)))))))))........((((------(.....)))))))) ( -52.00, z-score = -2.99, R) >droAna3.scaffold_13266 7128055 107 - 19884421 UGCGCCCCAGCUGAAGCGGAACGUCCACGCGCUUCUGGCUCCAGUUGGAGCUGAGAGUGUGGAAAGGAUUGGGGAUGGUGCUUAGGAGAGGGG------GAAUGUCGCCCGUA .(((((((..((.(((((...(((((.(((((((..((((((....))))))..)))))))...........))))).)))))...))..)))------).....)))..... ( -42.70, z-score = -1.11, R) >droWil1.scaffold_181009 1702788 83 + 3585778 UGAGCUCCCGCCGAGGCAGAGCGUCCGCGUGGAAGCGGCUCAAGAUUAUGUUUUUGUUGUGGAGCCGG-CGAAUGCCGUCCAUA----------------------------- (((((((((((.((.((...)).)).))).)))....)))))...............((((((..(((-(....))))))))))----------------------------- ( -29.10, z-score = -0.52, R) >droVir3.scaffold_13324 2669884 107 + 2960039 UGCGCACCCGCCGAGGAAGAGCGGCUUCGCUGCUGCUGU---GGCUCGAG-UGGAGGCGGUGCCAGG--UGAGAGGUGUGCAAGGGCGAAGGCUUAGGCGAAUAGCGCCCAUA (((((((((((((((..(.((((((......)))))).)---..)))).)-))..(((...)))...--.....)))))))).(((((...((....))......)))))... ( -48.30, z-score = -0.91, R) >droMoj3.scaffold_6496 12734587 104 - 26866924 UGCGCCCCCGCCGAGGCAGAGCGACUGCGCUGCUGCGGCUCAAAGGCGGG-CGCGGGCGGUGCCAGG--UGCGAACUGUGCCCAGGUGAAGGG------GAAUGGCGACCGUA .((((((((((((.(((((.((....)).))))).)))).....)).)))-)))..(((((((((((--(....)))...(((.......)))------...)))).))))). ( -51.40, z-score = -0.27, R) >consensus UGCGCACCAGCCGAGGCUGAGCGCCCACGAGUCUGCGGCUCCAGUUGGAGUUGCGAGUGGGGAAAGGACUGGGGUUGGUGCUUAGGCGAGGGC______GAAUACCGCCCGUA .((((....((....))......(((((.....(((((((((....))))))))).)))))................))))................................ (-18.80 = -18.84 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:34:02 2011