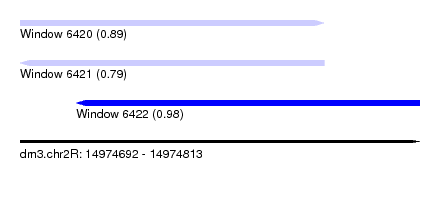
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,974,692 – 14,974,813 |
| Length | 121 |
| Max. P | 0.980528 |

| Location | 14,974,692 – 14,974,784 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.50350 |
| G+C content | 0.44418 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
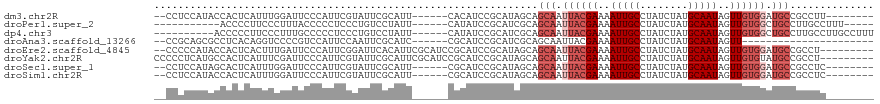
>dm3.chr2R 14974692 92 + 21146708 --------------------GAACUAUUUUGCAGUAAAAGGCG--GCAUCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGUGAAUGCGAAUACG------ --------------------.........(((........)))--((((.((((.((...((((((...(((((((.....))))))).)))))))).)))).)))).......------ ( -26.50, z-score = -2.14, R) >droPer1.super_2 6249015 109 + 9036312 UAACUAUUUAUCCCUGGCAGAAGGCGGCAAGAAAGGCAAGGCA-----GCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCGAUGCGGAUAUGAAUAGGACAGGG------ ...((((((((..(((.((....(((((((....(((......-----)))((....(((((........)))))....))..)))))))..)))))..)))))))).......------ ( -29.40, z-score = -1.22, R) >dp4.chr3 6047817 114 + 19779522 UAACUAUUUAUCCCUGGCAGAAGGCGGCAAGAAAGGCAAGGCAAGGCAGCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCGAUGCGGAUAUGAAUAGGACAGGG------ ...((((((((..(((.((....(((((((((((.((..(((......)))....(((((.....)))))))...))))....)))))))..)))))..)))))))).......------ ( -30.80, z-score = -1.11, R) >droAna3.scaffold_13266 2553800 77 - 19884421 ----------------------------------AACUAUACC--ACA-CCAAAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCGAUGCGGAUGCGGAUGCGAAUUGG------ ----------------------------------.........--...-((((..(((((.....)))))(((...((((((.((((......)))).)))))))))...))))------ ( -17.50, z-score = -0.99, R) >droEre2.scaffold_4845 9172544 97 + 22589142 --------------------GAACUAUUUCGCAGUAA-AGGCG--GCAUCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGCGGAUGCGAAUGUGAAUCCG --------------------.......((((((((..-..)).--((((((.((.((...((((((...(((((((.....))))))).)))))))).)).))))))...)))))).... ( -33.30, z-score = -2.92, R) >droYak2.chr2R 6932043 97 - 21139217 --------------------GAACUAUUUCGCAGUAA-AGGCG--GCAUACACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGCGGAUGCGAAUGCGAAUACG --------------------.......((((((....-...(.--((((.........((((((((...(((((((.....))))))).)))))))))))).).......)))))).... ( -28.74, z-score = -1.96, R) >droSec1.super_1 12480749 92 + 14215200 --------------------GAACUAUUUCGCAGUAAGAGGCG--GCAUCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGCGAAUGCGAAUACG------ --------------------.......((((((((.....)).--((((((.........((((((...(((((((.....))))))).))))))))))))...))))))....------ ( -30.60, z-score = -3.07, R) >droSim1.chr2R 13675823 92 + 19596830 --------------------GAACUAUUUCGCAGUAAGAGGCG--GCAUCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGCGAAUGCGAAUACG------ --------------------.......((((((((.....)).--((((((.........((((((...(((((((.....))))))).))))))))))))...))))))....------ ( -30.60, z-score = -3.07, R) >consensus ____________________GAACUAUUUCGCAGUAAAAGGCG__GCAUCCACAACUAUUGCAUAGAUAGGCAAUUUUCGUAAUUGCUGCUAUGCGGAUGCGAAUGCGAAUGCG______ ..........................................................((((((.(...(((((((.....))))))).).))))))....................... (-12.50 = -12.50 + 0.00)

| Location | 14,974,692 – 14,974,784 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.50350 |
| G+C content | 0.44418 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14974692 92 - 21146708 ------CGUAUUCGCAUUCACAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGC--CGCCUUUUACUGCAAAAUAGUUC-------------------- ------.......(((((((((...((((((..((((((.....))))))....))))))......)))))))))--.((........))..........-------------------- ( -23.90, z-score = -2.42, R) >droPer1.super_2 6249015 109 - 9036312 ------CCCUGUCCUAUUCAUAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGC-----UGCCUUGCCUUUCUUGCCGCCUUCUGCCAGGGAUAAAUAGUUA ------.......(((((..((((((((..(((((...(((((..(((((........)))))..)))))))-----)))..)))....((.((........)).))))))))))))... ( -28.60, z-score = -2.05, R) >dp4.chr3 6047817 114 - 19779522 ------CCCUGUCCUAUUCAUAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGCUGCCUUGCCUUGCCUUUCUUGCCGCCUUCUGCCAGGGAUAAAUAGUUA ------.......(((((..((((((((..(((((...(((((..(((((........)))))..))))))))))..))).........((.((........)).))))))))))))... ( -28.60, z-score = -1.56, R) >droAna3.scaffold_13266 2553800 77 + 19884421 ------CCAAUUCGCAUCCGCAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUUUGG-UGU--GGUAUAGUU---------------------------------- ------.......((....))..(((((((...((((((.....))))))((((.......))))....))-)))--)).......---------------------------------- ( -15.30, z-score = -0.21, R) >droEre2.scaffold_4845 9172544 97 - 22589142 CGGAUUCACAUUCGCAUCCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGC--CGCCU-UUACUGCGAAAUAGUUC-------------------- .((((....))))(((((((((...((((((..((((((.....))))))....))))))......)))))))))--(((..-.....))).........-------------------- ( -28.30, z-score = -2.48, R) >droYak2.chr2R 6932043 97 + 21139217 CGUAUUCGCAUUCGCAUCCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGUAUGC--CGCCU-UUACUGCGAAAUAGUUC-------------------- ....((((((...((((.((((...((((((..((((((.....))))))....))))))......)))).))))--.(...-...))))))).......-------------------- ( -23.60, z-score = -1.39, R) >droSec1.super_1 12480749 92 - 14215200 ------CGUAUUCGCAUUCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGC--CGCCUCUUACUGCGAAAUAGUUC-------------------- ------.......(((((((((...((((((..((((((.....))))))....))))))......)))))))))--(((........))).........-------------------- ( -25.30, z-score = -2.01, R) >droSim1.chr2R 13675823 92 - 19596830 ------CGUAUUCGCAUUCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGC--CGCCUCUUACUGCGAAAUAGUUC-------------------- ------.......(((((((((...((((((..((((((.....))))))....))))))......)))))))))--(((........))).........-------------------- ( -25.30, z-score = -2.01, R) >consensus ______CGCAUUCGCAUUCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGC__CGCCUUUUACUGCGAAAUAGUUC____________________ .............(((.(((((...((((.(..((((((.....))))))....).))))......))))).)))............................................. (-11.35 = -11.39 + 0.03)


| Location | 14,974,709 – 14,974,813 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.50609 |
| G+C content | 0.47772 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -9.82 |
| Energy contribution | -10.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14974709 104 - 21146708 --CCUCCAUACCACUCAUUUGGAUUCCCAUUCGUAUUCGCAUU------CACAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGCCGCCUU-------- --..((((...........))))...............(((((------((((...((((((..((((((.....))))))....))))))......)))))))))......-------- ( -25.80, z-score = -3.12, R) >droPer1.super_2 6249046 98 - 9036312 -----------ACCCCUUCCCUUUACCCCCUCCCUGUCCUAUU------CAUAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGCUGCCUUGCCUUU----- -----------................................------.......(((..(((((...(((((..(((((........)))))..))))))))))..)))....----- ( -19.90, z-score = -4.20, R) >dp4.chr3 6047848 104 - 19779522 ----------ACCCCCUUCCCUUUGCCCCCUCCCUGUCCUAUU------CAUAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGCUGCCUUGCCUUGCCUUU ----------.................................------.......(((..(((((...(((((..(((((........)))))..))))))))))..)))......... ( -19.90, z-score = -3.03, R) >droAna3.scaffold_13266 2553816 90 + 19884421 --CCGCAGCGCCUCACAGGUCCCCGUCCAUUCCAAUUCGCAUC------CGCAUCCGCAUCGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUU---------------------- --..((((((.......((.......))..........((...------.))...)))......((((((.....)))))).......))).......---------------------- ( -13.70, z-score = 0.07, R) >droEre2.scaffold_4845 9172561 109 - 22589142 --CCCCCAUACCACUCACUUUGAUUCCCAUUCGGAUUCACAUUCGCAUCCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGCCGCCU--------- --..................(((.(((.....))).))).....(((((((((...((((((..((((((.....))))))....))))))......))))))))).....--------- ( -30.30, z-score = -4.15, R) >droYak2.chr2R 6932060 111 + 21139217 CCCCCUCAUGCCACUCAUUUCGAUUCCCAUUCGUAUUCGCAUUCGCAUCCGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGUAUGCCGCCU--------- .......((((.........(((.......))).....))))..((((.((((...((((((..((((((.....))))))....))))))......)))).)))).....--------- ( -21.34, z-score = -1.65, R) >droSec1.super_1 12480766 104 - 14215200 --CCUCCAUAGCACUCAUUUGGAUUCCCAUUCGUAUUCGCAUU------CGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGCCGCCUC-------- --..((((((((........((((.(..((.((....)).)).------.).))))((((((..((((((.....))))))....))))))....)))))))).........-------- ( -26.60, z-score = -2.30, R) >droSim1.chr2R 13675840 104 - 19596830 --CCUCCAUACCACUCAUUUGGAUUCCCAUUCGUAUUCGCAUU------CGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGCCGCCUC-------- --..((((...........))))...............(((((------((((...((((((..((((((.....))))))....))))))......)))))))))......-------- ( -25.40, z-score = -2.49, R) >consensus __CCUCCAUACCACUCAUUUCGAUUCCCAUUCGUAUUCGCAUU______CGCAUCCGCAUAGCAGCAAUUACGAAAAUUGCCUAUCUAUGCAAUAGUUGUGGAUGCCGCCUC________ ................................................................(((.((((((..(((((........)))))..)))))).))).............. ( -9.82 = -10.83 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:53 2011