
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,966,906 – 14,966,997 |
| Length | 91 |
| Max. P | 0.628995 |

| Location | 14,966,906 – 14,966,997 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.67520 |
| G+C content | 0.38371 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -6.75 |
| Energy contribution | -5.86 |
| Covariance contribution | -0.90 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14966906 91 - 21146708 GAUUAUUGUUUUUCUUUUCCUGGUGCCCGGCAAACUGCAAUUGUUGC--UCGAGCACUUGACUUGACAGAUUUAAUCGCAUUAUUAUGCAAUG (((((..((((.((...((..(((((.((((((.(.......)))))--.)).))))).))...)).)))).)))))((((....)))).... ( -23.10, z-score = -2.41, R) >droSim1.chr2R 13668063 91 - 19596830 GAUUAUUGUUUUUCUUUUCCUGGUGCCCGGCAAACUGCAAUUGUUGC--UCGAGCACUUGACUUGACAGAUUUAAUCGCAUUAUUAUGCAAUG (((((..((((.((...((..(((((.((((((.(.......)))))--.)).))))).))...)).)))).)))))((((....)))).... ( -23.10, z-score = -2.41, R) >droSec1.super_1 12473251 91 - 14215200 GAUUAUUGUUUUUCUUUUCCUGGUGCCCGGCAAACUGCAAUUGUUGC--UCGAGCACUUGACUUGACAGAUUUAAUCGCAUUAUUAUGCAAUG (((((..((((.((...((..(((((.((((((.(.......)))))--.)).))))).))...)).)))).)))))((((....)))).... ( -23.10, z-score = -2.41, R) >droYak2.chr2R 6924087 91 + 21139217 GAUUAUUGUUUUUUUUUCUCUGGCGACCGGCAAACUGCAAUUGUUGC--UCAAGCACUUGACUUGACAGUUUUAAUCGCAUUAUUAUGCAAUG (((((..............((((((((..((.....))....)))))--((((((....).))))))))...)))))((((....)))).... ( -17.83, z-score = -0.72, R) >droEre2.scaffold_4845 9164586 87 - 22589142 GAUUAUUGGUUUUU----UCUGGCAACCGGCGAGCUGCAAUUGUUGC--UCGAGCACUUGACUUGACAGUUUUAAUCGCAUUAUUAUGCAAUG .......((((...----.((((((((..((.....))....)))))--((((((....).))))))))....))))((((....)))).... ( -19.00, z-score = -0.19, R) >droAna3.scaffold_13266 2542708 82 + 19884421 ---------GAAUACGUAAAUGACUGUUUCAACUGUGCAACUGUUGC--UCGAGCACUUGACCUGGCCGUUUUAAUCGCAUUAUUAUGCGAGG ---------........(((((.(((..((((..((((.........--....))))))))..))).)))))...((((((....)))))).. ( -15.32, z-score = 0.47, R) >droWil1.scaffold_180697 2621906 83 - 4168966 --------GUUUUAGUAUUGUUUUUGCUGUCCAGUUGCAACUGUUGC--UUAAGCACUUGACUCGACAGUUUUAAUCACAUUAUUAUGCCCUG --------((..(((((.(((....((((((.(((..(((.((((..--...)))).)))))).)))))).......))).))))).)).... ( -17.00, z-score = -1.55, R) >droVir3.scaffold_12875 1141365 75 + 20611582 ------------------GUUUAUUGUUUUUGCUGUCCAACUGUUGCACUCGAGCACUUGACUCGACAGUUUUAAUCAUAUUAUUUUGUCUCC ------------------...(((.(((...(((((((((.((((.......)))).)))....))))))...))).)))............. ( -10.30, z-score = -0.35, R) >droGri2.scaffold_15245 18082025 75 - 18325388 ------------------GUUUUUAAUUUUUACUGACCAAUUGUUGCACUUGAGUGCUUUAAGCAACGGGUUUAAUCAUAUUAUUUUGUUUUC ------------------.....((((.......((((..(((((((((....))))....)))))..)))).......)))).......... ( -12.54, z-score = -1.30, R) >consensus GAUUAUUGUUUUUCUUUUCCUGGUGACCGGCAAACUGCAAUUGUUGC__UCGAGCACUUGACUUGACAGUUUUAAUCGCAUUAUUAUGCAAUG ........................................((((((...((((....))))..))))))........((((....)))).... ( -6.75 = -5.86 + -0.90)

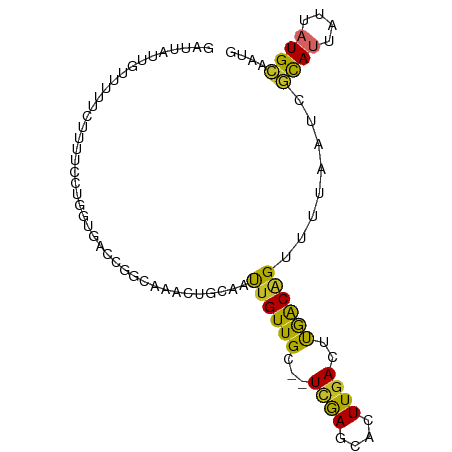
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:51 2011