
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,966,588 – 14,966,653 |
| Length | 65 |
| Max. P | 0.611832 |

| Location | 14,966,588 – 14,966,653 |
|---|---|
| Length | 65 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 69.33 |
| Shannon entropy | 0.68113 |
| G+C content | 0.45778 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -6.22 |
| Energy contribution | -6.05 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
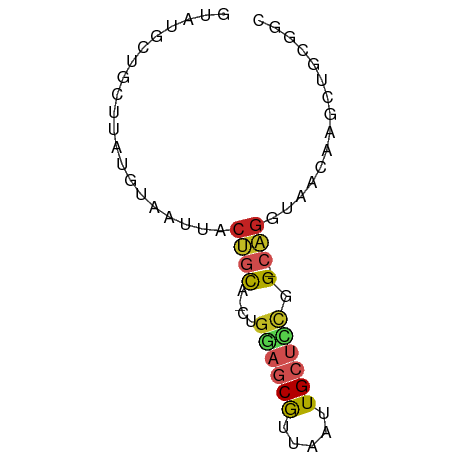
>dm3.chr2R 14966588 65 + 21146708 GUAUGCUGCUUAUGUAAUUACUGCUCCUGGAGCGUUAAUUGCUCCGG-CAGGUAACAAGCUGCGGC ....(((((...(((.....(((((...((((((.....))))))))-)))...)))....))))) ( -23.10, z-score = -1.91, R) >droSim1.chr2R 13667746 65 + 19596830 GUAUGCUGCUUAUGUAAUUACUGCUGCUGGAUCGUUAAUUGCUCCGG-CAGGUAACAAGCUGCGGC ....(((((....((..((((..((((((((.((.....)).)))))-)))))))...)).))))) ( -21.40, z-score = -1.64, R) >droSec1.super_1 12472936 65 + 14215200 GUAUGCAGCUUAUGUAAUUACUGCUGCUGGAGCGUUAAUUGCUCCGG-CAGGUAACAAGCUGCGGC ...(((((((..(((...(((..(((((((((((.....))))))))-)))))))))))))))).. ( -31.30, z-score = -4.39, R) >droYak2.chr2R 6923731 65 - 21139217 GUAUGCUGCUUAUGUAAUUACUACUGCUGGAGCGUUAAUUGCUCCGG-CAGGUAACAAGCUGCGGC ....(((((....((..((((..(((((((((((.....))))))))-)))))))...)).))))) ( -28.00, z-score = -3.88, R) >droEre2.scaffold_4845 9164246 65 + 22589142 GUGUGCUGCUUAUGUAAUUACUGCUGCUGGAGCGUUAAUUGCUCCGG-CAGGUAACAAGCUGCGGC ....(((((....((..((((..(((((((((((.....))))))))-)))))))...)).))))) ( -28.00, z-score = -3.14, R) >droAna3.scaffold_13266 2542400 66 - 19884421 GCAUAUUGCAUAUGUAAUUACUGCAUCUGGAGCGUUAAUUGCUCUGGACAGGUAACAAGCUGCCGC (((.(((((....)))))...))).((..(((((.....)))))..))..((((......)))).. ( -21.00, z-score = -1.90, R) >dp4.chr3 6039013 63 + 19779522 AUUUGUUGCAUAUGUAAUAGCUGCA---GAAGCGGUAAUUGCUUUGGCCAGGUAACAAGCAGGCAC .((((((((....(((((.(((((.---...))))).))))).........))))))))....... ( -16.12, z-score = -0.09, R) >droPer1.super_2 6240168 63 + 9036312 AUUUGUUGCAUAUGUAAUAGCUGCA---GAAGCGGUAAUUGCUUUGGCCAGGUAACAAGCAGGCAC .((((((((....(((((.(((((.---...))))).))))).........))))))))....... ( -16.12, z-score = -0.09, R) >droWil1.scaffold_180697 2621654 62 + 4168966 CAACCAUACAUAUAUAAUUACUGUG---GCUGCGUUAAUUGCUCUGGCCAGGUAA-ACGUAAAUGC ........(((.(((..(((((.((---((((((.....)))...))))))))))-..))).))). ( -13.10, z-score = -0.51, R) >droVir3.scaffold_12875 1141124 60 - 20611582 --AGGUAAUGCAUAUAAUUACGGCA---GCGGCGUUAAUUGCAGCAG-CUCGUUGAAUGCCACAUA --..(((((.......)))))((((---.(((((......((....)-).)))))..))))..... ( -13.80, z-score = 0.40, R) >droGri2.scaffold_15245 18081770 57 + 18325388 --AAGUAAUGCACAUAAUUACGCCA---ACAACAUUAAUUGCUUCAG-AAGGUAGUAUAUUUC--- --..(((((.......)))))....---.........((((((....-..)))))).......--- ( -6.20, z-score = -0.29, R) >consensus GUAUGCUGCUUAUGUAAUUACUGCA_CUGGAGCGUUAAUUGCUCCGG_CAGGUAACAAGCUGCGGC ...................(((......((((((.....)))))).....)))............. ( -6.22 = -6.05 + -0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:50 2011