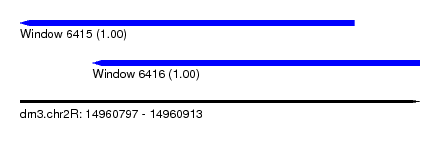
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,960,797 – 14,960,913 |
| Length | 116 |
| Max. P | 0.999656 |

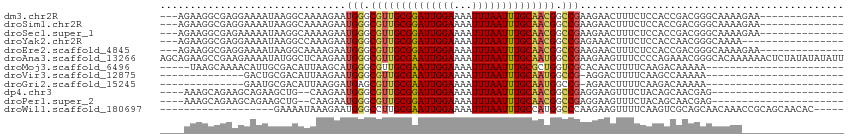
| Location | 14,960,797 – 14,960,894 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 66.42 |
| Shannon entropy | 0.64460 |
| G+C content | 0.43073 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -17.97 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998521 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 14960797 97 - 21146708 ---AGAAGGCGAGGAAAAUAAGGCAAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUUCUCCACCGACGGGCAAAAGAA-------------- ---.(..((.(..((((......(....)..(((.((((((((((((((...)))))))))))))).)))......))))..).))..)...........-------------- ( -24.80, z-score = -1.29, R) >droSim1.chr2R 13661854 97 - 19596830 ---AGAAGGCGAGGAAAAUAAGGCAAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUUCUCCACCGACGGGCAAAAGAA-------------- ---.(..((.(..((((......(....)..(((.((((((((((((((...)))))))))))))).)))......))))..).))..)...........-------------- ( -24.80, z-score = -1.29, R) >droSec1.super_1 12467112 97 - 14215200 ---AGAAGGCGAGAAAAAUAAGGCAAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUUCUCCACCGACGGGCAAAAGAA-------------- ---.(..((.((((((.......(....)..(((.((((((((((((((...)))))))))))))).)))......))))))..))..)...........-------------- ( -25.10, z-score = -1.56, R) >droYak2.chr2R 6916953 94 + 21139217 ---AGAAGGCGAGGAAAAUAAGGCCAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGAAACUUUCUCCACCAACGGGCAAAA----------------- ---...................(((.......((.((((((((((((((...)))))))))))))).))((((.....))))........)))....----------------- ( -28.10, z-score = -2.13, R) >droEre2.scaffold_4845 9158273 97 - 22589142 ---AGAAGGCGAGGAAAAUAAGGCAAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUUCUCCACCGACGGGCAAAAGAA-------------- ---.(..((.(..((((......(....)..(((.((((((((((((((...)))))))))))))).)))......))))..).))..)...........-------------- ( -24.80, z-score = -1.29, R) >droAna3.scaffold_13266 2535775 114 + 19884421 AGCAGAAGCCGAAGAAAAUAUGGCUCAAGAAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCGAAGAAGUUUCCCCAGAAACGGGCACAAAAAACUCUAUAUAUAUU .((....))...........((((((.....(((.((((((((((((((...)))))))))))))).))).....(((((....))))))))).)).................. ( -29.40, z-score = -2.00, R) >droMoj3.scaffold_6496 20362363 86 + 26866924 -----UAAGCAAAACAUUGCGACAUUAAGCAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCGCUGGUCGCACAACUUUUCAAGACAAAAA----------------------- -----............((((((.....((....))...((((((((((...))))))))))....))))))...................----------------------- ( -18.20, z-score = -0.42, R) >droVir3.scaffold_12875 1133623 76 + 20611582 --------------GACUGCGACAUUAAGAAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCG-AGGACUUUUCAAGCCAAAAA----------------------- --------------.................(((.((((((((((((((...)))))))))))))).)))-.((.((.....)))).....----------------------- ( -20.40, z-score = -2.08, R) >droGri2.scaffold_15245 18066959 76 - 18325388 --------------GAAUGCGACAUUAAGGAUGAGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCG-AGAACUUUUCAAGACAAAAA----------------------- --------------....((..((((...))))..((((((((((((((...)))))))))))))))).(-(((...))))..........----------------------- ( -14.20, z-score = -0.81, R) >dp4.chr3 6032924 86 - 19779522 ----AAAGCAGAAGCAGAAGCUG--CAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGGAAGUUUCUACAGCAACGAG---------------------- ----...((....))(((((((.--(.....(((.((((((((((((((...)))))))))))))).)))..).)))))))...........---------------------- ( -27.50, z-score = -3.02, R) >droPer1.super_2 6234042 86 - 9036312 ----AAAGCAGAAGCAGAAGCUG--CAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGGAAGUUUCUACAGCAACGAG---------------------- ----...((....))(((((((.--(.....(((.((((((((((((((...)))))))))))))).)))..).)))))))...........---------------------- ( -27.50, z-score = -3.02, R) >droWil1.scaffold_180697 2611975 90 - 4168966 -------------------GAAAAUAAAGAAUGGGCCUUGCGAAUUGGAAAAUUUAAUUUGCCAUGGCCCAAGAAGUUUUCAAGUCGCAGCAACAAACCGCAGCAACAC----- -------------------((((((......((((((..((((((((((...))))))))))...))))))....))))))..((.((.((........)).)).))..----- ( -27.30, z-score = -3.34, R) >consensus ____GAAGGCGAGGAAAAUAAGGCUAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUUCUACACCAACGGGCAAAA_________________ ................................((.((((((((((((((...)))))))))))))).))............................................. (-17.97 = -17.73 + -0.24)

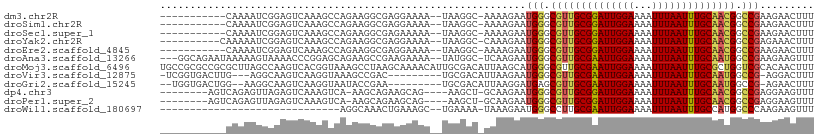
| Location | 14,960,818 – 14,960,913 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.51 |
| Shannon entropy | 0.67328 |
| G+C content | 0.43223 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -17.97 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14960818 95 - 21146708 -----------CAAAAUCGGAGUCAAAGCCAGAAGGCGAGGAAAA--UAAGGC-AAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUU -----------.......((((((...(((....)))........--......-.......(((.((((((((((((((...)))))))))))))).)))..).))))) ( -26.50, z-score = -3.05, R) >droSim1.chr2R 13661875 95 - 19596830 -----------CAAAAUCGGAGUCAAAGCCAGAAGGCGAGGAAAA--UAAGGC-AAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUU -----------.......((((((...(((....)))........--......-.......(((.((((((((((((((...)))))))))))))).)))..).))))) ( -26.50, z-score = -3.05, R) >droSec1.super_1 12467133 95 - 14215200 -----------CAAAAUCGGAGUCAAAGCCAGAAGGCGAGAAAAA--UAAGGC-AAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUU -----------.......((((((...(((....)))........--......-.......(((.((((((((((((((...)))))))))))))).)))..).))))) ( -26.50, z-score = -3.18, R) >droYak2.chr2R 6916971 96 + 21139217 ----------CAAAAAUCGGAGUCAAAGCCAGAAGGCGAGGAAAA--UAAGGC-CAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGAAACUUU ----------......((((..((...(((....)))...))...--....((-(........)))(((((((((((((...)))))))))))))..))))........ ( -27.30, z-score = -2.71, R) >droEre2.scaffold_4845 9158294 95 - 22589142 -----------CAAAAUCGGAGUCAAAGCCAGAAGGCGAGGAAAA--UAAGGC-AAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUU -----------.......((((((...(((....)))........--......-.......(((.((((((((((((((...)))))))))))))).)))..).))))) ( -26.50, z-score = -3.05, R) >droAna3.scaffold_13266 2535810 103 + 19884421 ---GGCAGAAUAAAAAGUAAAACCCGGAGCAGAAGCCGAAGAAAA--UAUGGC-UCAAGAAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCGAAGAAGUUU ---(((................((((...(...(((((.(.....--).))))-)...)..))))((((((((((((((...))))))))))))))))).......... ( -27.80, z-score = -2.80, R) >droMoj3.scaffold_6496 20362375 109 + 26866924 UGCCGCGCCGCGCUUAGCCAAGUCACGGUAAAGCCUAAGCAAAACAUUGCGACAUUAAGCAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCGCUGGUCGCACAACUUU ....((((((((((((((...(((..((.....))...((((....))))))).....)).))))))).((((((((((...))))))))))...)).)))........ ( -31.70, z-score = 0.25, R) >droVir3.scaffold_12875 1133635 95 + 20611582 -UCGGUGACUUG---AGGCAAGUCAAGGUAAAGCCGAC---------UGCGACAUUAAGAAUGGGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCG-AGGACUUU -(((((..((((---(......))))).....))))).---------..............(((.((((((((((((((...)))))))))))))).)))-........ ( -29.50, z-score = -2.64, R) >droGri2.scaffold_15245 18066971 95 - 18325388 --UGGUGACUGG--AAGGCAAGUCAAGGUAAUACCGAA---------UGCGACAUUAAGGAUGAGCGUUGCGAAUUGGAAAAUUUAAUUUGCAAUGGCCG-AGAACUUU --.(....)...--..(((..(((..((.....))(..---------..))))............((((((((((((((...))))))))))))))))).-........ ( -22.70, z-score = -1.45, R) >dp4.chr3 6032937 95 - 19779522 --------AGUCAGAGUUAGAGUCAAAGUCA-AAGCAGAAGCAG----AAGCU-GCAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGGAAGUUU --------.....((.((.......)).)).-((((....((((----...))-)).....(((.((((((((((((((...)))))))))))))).))).....)))) ( -25.30, z-score = -2.08, R) >droPer1.super_2 6234055 95 - 9036312 --------AGUCAGAGUUAGAGUCAAAGUCA-AAGCAGAAGCAG----AAGCU-GCAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAGGAAGUUU --------.....((.((.......)).)).-((((....((((----...))-)).....(((.((((((((((((((...)))))))))))))).))).....)))) ( -25.30, z-score = -2.08, R) >droWil1.scaffold_180697 2612005 76 - 4168966 ------------------------------AGGCAAACUGAAAGC--UGAAAA-UAAAGAAUGGGCCUUGCGAAUUGGAAAAUUUAAUUUGCCAUGGCCCAAGAAGUUU ------------------------------.(((.........))--).....-.......((((((..((((((((((...))))))))))...))))))........ ( -21.10, z-score = -2.35, R) >consensus ___________CAAAAUCGGAGUCAAAGCCAGAAGGAGAGGAAAA__UAAGGC_UAAAGAAUGGGCGUUGCGGAUUGGAAAAUUUAAUUUGCAACGGCCGAAGAACUUU ..............................................................((.((((((((((((((...)))))))))))))).)).......... (-17.97 = -17.73 + -0.24)
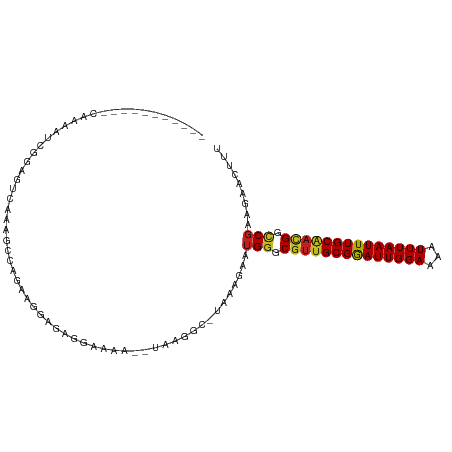
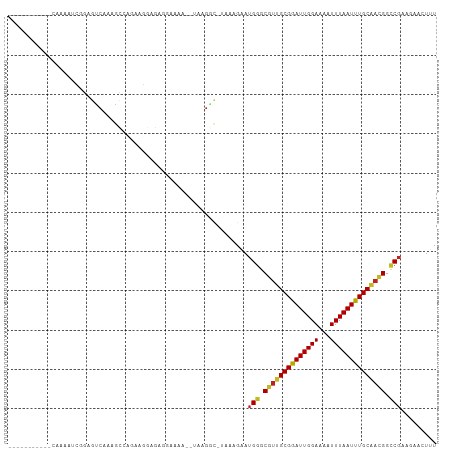
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:48 2011