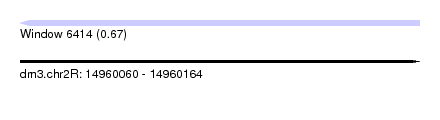
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,960,060 – 14,960,164 |
| Length | 104 |
| Max. P | 0.666069 |

| Location | 14,960,060 – 14,960,164 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.77 |
| Shannon entropy | 0.56208 |
| G+C content | 0.54492 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.49 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
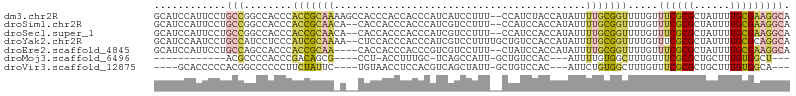
>dm3.chr2R 14960060 104 - 21146708 GCAUCCAUUCCUGCCGGCCACCCACCGCAAAAGCCACCCACCACCCAUCAUCCUUU--CCAUCUACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGAAGGCA (((........)))..(((....(((((((((........................--............))))))))).....((((((......))))))))). ( -21.72, z-score = -1.99, R) >droSim1.chr2R 13661139 102 - 19596830 GCAUCCAUUCCUGCCGGCCACCCACCGCAACA--CACCACCCACCCAUCGUCCUUU--CCAUCCACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGAAGGCA (((........)))..(((....(((((((..--......................--..............))))))).....((((((......))))))))). ( -18.96, z-score = -1.03, R) >droSec1.super_1 12466393 102 - 14215200 GCAUCCAUUCCUGCCGGCCACCCACCGCAACA--CACCACCCACCCAUCGUCCUUU--CCAUCCACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGAAGGCA (((........)))..(((....(((((((..--......................--..............))))))).....((((((......))))))))). ( -18.96, z-score = -1.03, R) >droYak2.chr2R 6916229 104 + 21139217 GCAUCCAAUCCUGCCAUCCUCCCAUCGCAAAA--CUCCACCCACCCAUCGUCCUUUUGCUGUCCACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGCAGGCA ............(((........(((((((((--...........((.((......)).)).........)))))))))........((((......)))).))). ( -20.05, z-score = -1.92, R) >droEre2.scaffold_4845 9157564 100 - 22589142 GCAUCCAUUCCUGCCAGCCACCCACCGCAA----CACCACCCACCCGUCGUCCUUU--CUAUCCACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGAAGGCA (((........)))..(((....(((((((----......................--..............))))))).....((((((......))))))))). ( -19.22, z-score = -1.87, R) >droMoj3.scaffold_6496 20361548 81 + 26866924 ------------ACGCCCCACCCGACAGCG----CCU-ACCUUUGC-UCAGCCAUU-GCUGUCCAC---AUUUUGUGGCUUUGUUUCGCGCUGCUUUGUGGCU--- ------------..(((.((...(.(((((----(..-........-.((((....-)))).((((---.....)))).........)))))))..)).))).--- ( -24.10, z-score = -1.78, R) >droVir3.scaffold_12875 1132941 91 + 20611582 ----GCACCCCCACGGCCCCCCUUCUAUUC----UGUAACCUCCACGUCAGCUAUU-GCUGUCCAC---AUUCUGUGGCUUUGUUUCGCGCUGCUUUGUGGCA--- ----(((...((((((..............----.((.......))(.((((....-)))).)...---...))))))...))).....((..(...)..)).--- ( -16.70, z-score = 0.40, R) >consensus GCAUCCAUUCCUGCCGGCCACCCACCGCAA_A__CACCACCCACCCAUCGUCCUUU__CUAUCCACCAUAUUUUGCGGUUUUGUUUCGCGCUAUUUUGCGAAGGCA ............(((........(((((((..........................................))))))).....((((((......))))))))). ( -9.10 = -9.49 + 0.40)
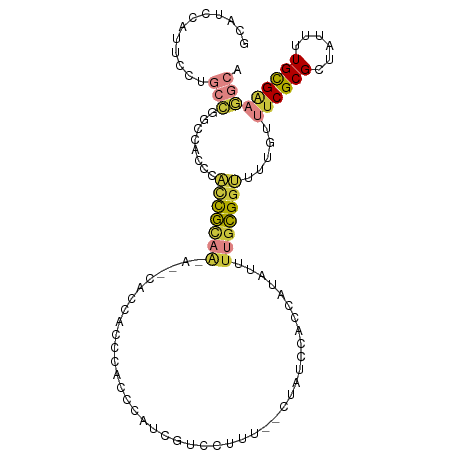
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:47 2011