| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,246,431 – 3,246,524 |
| Length | 93 |
| Max. P | 0.910767 |

| Location | 3,246,431 – 3,246,524 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.31320 |
| G+C content | 0.48838 |
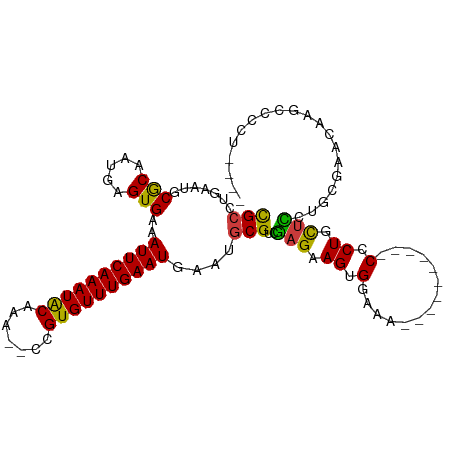
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -13.79 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910767 |
| Prediction | RNA |
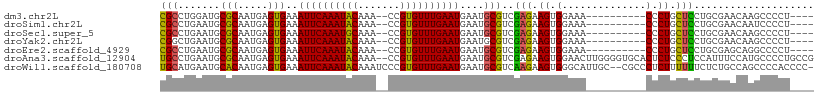
Download alignment: ClustalW | MAF
>dm3.chr2L 3246431 93 + 23011544 CGCCUGGAUGCGCAAUGAGUGAAAUUCAAAUACAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA----------CCCUGCUCCUGCGAACAAGCCCCU---- .((.((....((((..(((((..((((((((((...--..))))))))))...))).))(((.((.(....----------).)).))).))))..)).))....---- ( -25.10, z-score = -2.58, R) >droSim1.chr2L 3190194 93 + 22036055 CGCCUGAAUGCGCAAUGAGUGAAAUUCAAAUACAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA----------CCCUGCUCCUGCGAACAAUCCCCU---- (((......(((((.........((((((((((...--..))))))))))...))))).(((.((.(....----------).)).)))..)))...........---- ( -22.00, z-score = -2.54, R) >droSec1.super_5 1395948 93 + 5866729 CGCCUGAAUGCGCAAUGAGUGAAAUUCAAAUGCAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA----------CCCUGCUCCUGCGAACAAGCCCCU---- .((.((....((((..(((((..((((((((((...--..))))))))))...))).))(((.((.(....----------).)).))).))))..)).))....---- ( -23.00, z-score = -2.13, R) >droYak2.chr2L 3236263 93 + 22324452 CGGCUGAAUGCGCAAUGAGUGAAAUUCAAAUACAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA----------CCCUGCUCCUGCGAACAAGCCCCU---- .((((.....((((..(((((..((((((((((...--..))))))))))...))).))(((.((.(....----------).)).))).))))....))))...---- ( -28.90, z-score = -3.93, R) >droEre2.scaffold_4929 3283775 93 + 26641161 CGCCUGAAUGCGCAAUGAGUGAAAUUCAAAUACAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA----------CCCUGCUCCUGCGAGCAGGCCCCU---- .(((((....((((..(((((..((((((((((...--..))))))))))...))).))(((.((.(....----------).)).))).))))..)))))....---- ( -31.60, z-score = -4.35, R) >droAna3.scaffold_12904 56087 107 - 61084 UGCCUGAAUGCGCAAUGAGUGAAAUUCAAAUACAAA--CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAACUUGGGGUGCACUCUCCCUCCAUUUCCAUGCCCCUGCCG .((......))(((.........((((((((((...--..))))))))))....((((.(.((((((((....((((......)))).)))))))))))))...))).. ( -29.10, z-score = -1.61, R) >droWil1.scaffold_180708 504659 106 - 12563649 UGCAUGAAUGCACAAUGAGUGAAAUUCAAAUACAAAUCCCGUGUUUGAAUGAAUGCGUCAAGAAGUGGGCAUUGC--CGCCCUCUUUUUUCUCUGCCAGCCCCACCCC- .((((.....(((.....)))..((((((((((.......))))))))))..))))...((((((.((((.....--.)))).))))))...................- ( -26.80, z-score = -1.98, R) >consensus CGCCUGAAUGCGCAAUGAGUGAAAUUCAAAUACAAA__CCGUGUUUGAAUGAAUGCGUCGAGAAGUGGAAA__________CCCUGCUCCUGCGAACAAGCCCCU____ (((.......(((.....)))..((((((((((.......))))))))))....)))..(((.((.((.............)))).))).................... (-13.79 = -12.98 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:25 2011