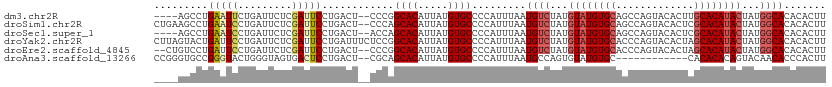
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,949,059 – 14,949,165 |
| Length | 106 |
| Max. P | 0.816420 |

| Location | 14,949,059 – 14,949,165 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.34713 |
| G+C content | 0.46694 |
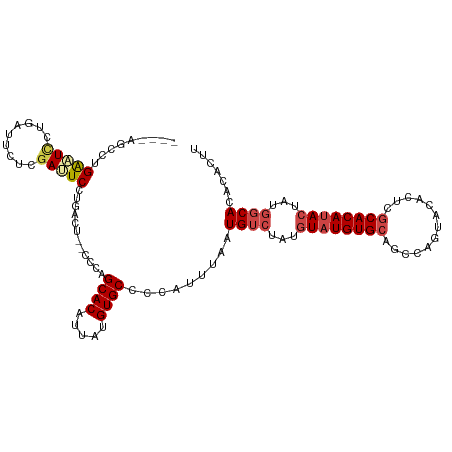
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
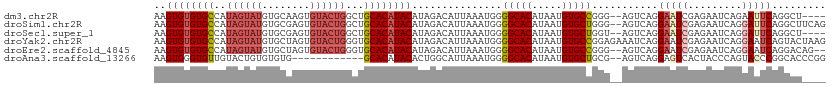
>dm3.chr2R 14949059 106 + 21146708 AAGUGUGUGCCAUAGUAUGUGCAAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGGG--AGUCAGGAAUCGAGAAUCAGAAUUCAGGCU---- ...((((((((...(((((((((.((......))))))))))).....(((...))).))))))))....(((..(--(((...((........))...)))).))).---- ( -33.40, z-score = -1.96, R) >droSim1.chr2R 13648579 110 + 19596830 AAGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCUGGG--AGUCAGGAAUCGAGAAUCAGGAUUCAGGCUUCAG ...((((((((...((((((((.(((......))))))))))).....(((...))).))))))))........((--((((..(((((.........))))).)))))).. ( -37.00, z-score = -2.51, R) >droSec1.super_1 12455528 106 + 14215200 AAGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCUGGU--AGUCAGGAAUCGAGAAUCAGGAUUCAGGCU---- ...((((((((...((((((((.(((......))))))))))).....(((...))).))))))))....(((.(.--((((..((........))..))))).))).---- ( -32.40, z-score = -1.29, R) >droYak2.chr2R 6904726 112 - 21139217 AAGUGUGUGCCAUAGUAUGUGCUAGUGUACUGGGUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGGAGAAAUCAGGAAUCGAGAAUCAGGAAUCAGUACUAAG ..(((((..((.(((((..(....)..)))))))..)))))...(((((.........(((((.....))))).((....))......................)).))).. ( -31.30, z-score = -1.79, R) >droEre2.scaffold_4845 9146208 108 + 22589142 AAGUGUGUGCCAUAGUAUGUGCUAGUGUACUGGGUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGGG--AGUCAGGAAUCGAGAAUCAGGAAUCAGGACAG-- ..(((((..((.(((((..(....)..)))))))..))))).................(((((.....)))))...--.(((..((.((.........)).))..)))..-- ( -33.10, z-score = -2.04, R) >droAna3.scaffold_13266 2523024 98 - 19884421 AAGUGGGUGUUGUACUGUGUGUG------------GCACAUACACUGGCAUUAAAUGGGGCACAUAAUGUGCUGCG--AGUCAGGAGUCACUACCCAGUACCCGGCACCCGG ...((((((((((((((.(((((------------((.......(((((......((.(((((.....))))).))--.)))))..))))).)).))))))..)))))))). ( -38.10, z-score = -1.10, R) >consensus AAGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGGG__AGUCAGGAAUCGAGAAUCAGGAUUCAGGAC____ ..((((((((..(((((..(....)..)))))...))))))))...............(((((.....)))))...........(((((.........)))))......... (-22.84 = -24.28 + 1.45)
| Location | 14,949,059 – 14,949,165 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.34713 |
| G+C content | 0.46694 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
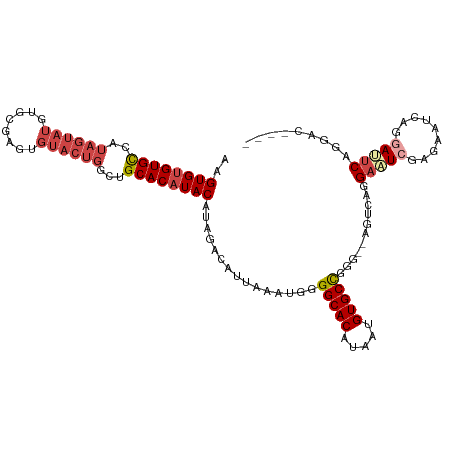
>dm3.chr2R 14949059 106 - 21146708 ----AGCCUGAAUUCUGAUUCUCGAUUCCUGACU--CCCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUUGCACAUACUAUGGCACACACUU ----.(((.((((....)))).........(((.--...(((((.....))))).........)))...((((((((((.........))))))))))...)))........ ( -25.82, z-score = -1.78, R) >droSim1.chr2R 13648579 110 - 19596830 CUGAAGCCUGAAUCCUGAUUCUCGAUUCCUGACU--CCCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACUU ..(((....((((....))))....)))......--....((((.....)))).........((((...((((((((((........)).))))))))...))))....... ( -23.90, z-score = -1.26, R) >droSec1.super_1 12455528 106 - 14215200 ----AGCCUGAAUCCUGAUUCUCGAUUCCUGACU--ACCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACUU ----.(((.(((((.........)))))..(((.--....((((.....))))..........)))...((((((((((........)).))))))))...)))........ ( -23.36, z-score = -1.27, R) >droYak2.chr2R 6904726 112 + 21139217 CUUAGUACUGAUUCCUGAUUCUCGAUUCCUGAUUUCUCCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCACCCAGUACACUAGCACAUACUAUGGCACACACUU .......................................(((((.....)))))........((((...((((((((.............))))))))...))))....... ( -23.22, z-score = -1.35, R) >droEre2.scaffold_4845 9146208 108 - 22589142 --CUGUCCUGAUUCCUGAUUCUCGAUUCCUGACU--CCCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCACCCAGUACACUAGCACAUACUAUGGCACACACUU --..(((..((.((.........)).))..))).--...(((((.....)))))........((((...((((((((.............))))))))...))))....... ( -25.72, z-score = -2.16, R) >droAna3.scaffold_13266 2523024 98 + 19884421 CCGGGUGCCGGGUACUGGGUAGUGACUCCUGACU--CGCAGCACAUUAUGUGCCCCAUUUAAUGCCAGUGUAUGUGC------------CACACACAGUACAACACCCACUU ..(((((....((((((((((((((........)--))).((((.....)))).........)))).((((......------------.)))).))))))..))))).... ( -33.10, z-score = -0.85, R) >consensus ____AGCCUGAAUCCUGAUUCUCGAUUCCUGACU__CCCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACUU .........(((((.........)))))............((((.....)))).........((((...((((((((.............))))))))...))))....... (-17.03 = -17.95 + 0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:45 2011