
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,947,998 – 14,948,091 |
| Length | 93 |
| Max. P | 0.877207 |

| Location | 14,947,998 – 14,948,091 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 54.63 |
| Shannon entropy | 0.88788 |
| G+C content | 0.52176 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -9.45 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
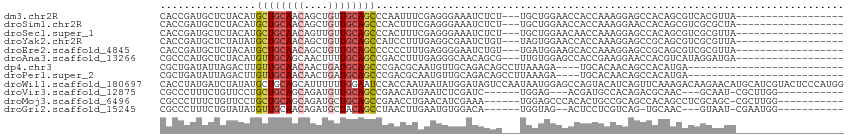
>dm3.chr2R 14947998 93 + 21146708 CACCGAUGCUCUACAUGCUGCAACAGCUGUUGCAGCCCAAUUUCGAGGGAAAUCUCU---UGCUGGAACCACCAAAGGAGCCACAGCGUCACGUUA------------------ ....((((((......((((((((....)))))))).......((((((....))))---)).(((..((......))..))).))))))......------------------ ( -31.00, z-score = -2.28, R) >droSim1.chr2R 13647541 93 + 19596830 CACCGAUGCUCUACAUGCUGCAACAGCUGUUGCAGCCCACUUUCGAGGGAAAUCUCU---UGCUGGAACCACCAAAGGAACCACAGCGUCGCGCUA------------------ ...(((((((......((((((((....))))))))((.....((((((....))))---)).(((.....)))..))......))))))).....------------------ ( -31.00, z-score = -2.32, R) >droSec1.super_1 12454498 93 + 14215200 CACCGAUGCUCUACAUGCUGCAACAGUUGUUGCAGCCCACUUUCGAGGGAAAUCUCU---UGCUGGAACAACCAAAGGAGCCACAGCGUCGCGUUA------------------ .(((((((((......((((((((....))))))))((.....((((((....))))---)).(((.....)))..))......))))))).))..------------------ ( -31.90, z-score = -2.27, R) >droYak2.chr2R 6903685 93 - 21139217 CACCGAUGCUCUAUAUGCUGCAACAGCUGUUGCAGCCAUCCUUUGAGGCGAAUCUGU---UAGUGGAACCACCAAAGGAGCCGCAGCGUCGCGUUA------------------ .((((((((((.....((((((((....))))))))..(((((((.((((....)))---).(((....))))))))))...).))))))).))..------------------ ( -34.90, z-score = -2.50, R) >droEre2.scaffold_4845 9145170 93 + 22589142 CACCGAUGCUCUACAUGCUGCAACAGCUGUUGCAGCCCCCCUUUGAGGGGAAUCUGU---UGAUGGAAGCACCAAAGGAGCCGCAGCGUCGCGUUA------------------ .((((((((((.....((((((((....)))))))).((((.....))))..(((.(---((.((....)).))).)))...).))))))).))..------------------ ( -35.10, z-score = -1.78, R) >droAna3.scaffold_13266 2521008 93 - 19884421 CGCCCAUGCUCUACAUGUUGCAGCAACUUUUGCAGCCGACCUUUGAGGGCAACAGCG---UUGUGGAGCCACCGAAGGAACCACGUCAUAGGAUGA------------------ ....(((.((..((..(((((((......)))))))...((((((..(((..((...---...))..)))..))))))......))...)).))).------------------ ( -26.00, z-score = 0.10, R) >dp4.chr3 6021550 84 + 19779522 CGCUGAUAUUAGACUUGUUGCAACAACUGAUGCAGCCCGACGCAAUGUUGCAGACAGCCUUAAAGA----UGCACAACAGCCACAUGA-------------------------- .((((..........((((((((((.....(((........))).))))))).)))((........----.))....)))).......-------------------------- ( -16.10, z-score = 0.41, R) >droPer1.super_2 6222608 84 + 9036312 CGCUGAUAUUAGACUUGUUGCAACAACUGAUGCAGCCCGACGCAAUGUUGCAGACAGCCUUAAAGA----UGCACAACAGCCACAUGA-------------------------- .((((..........((((((((((.....(((........))).))))))).)))((........----.))....)))).......-------------------------- ( -16.10, z-score = 0.41, R) >droWil1.scaffold_180697 2593757 114 + 4168966 CACCUAUGAUCUAUAUGCUGCAGCAUUUUUUGGAAUCCACCAAUAAUAUGGAUAGUCCAAUAAUGGAGCCAGUACAUCAGUUCAAAGACAAGAACAUGCAUCGUACUCCCAUGG ...(((((.(((((((((....))))...((((((((((.........)))))..)))))..)))))...(((((((((((((........)))).)).)).)))))..))))) ( -24.30, z-score = -0.80, R) >droVir3.scaffold_12875 1117847 90 - 20611582 CGCCCUUUCUGUUCCUGCUGCAGCAGAUGUUGCAGCCGAACAUGAAUCUCGAUC------UGGAG---ACGAUGCCACAGACGCAAC---GCAAU-CGCUUGG----------- .((...((((((((..((((((((....)))))))).))))).)))((((....------..)))---)((.(((.......))).)---)....-.))....----------- ( -26.80, z-score = -0.67, R) >droMoj3.scaffold_6496 20341079 96 - 26866924 CGCCCUUUCUGUUCCUGCUGCAGCAGAUGCUGCAGCCGAACCUGAACAUCGAAA------UGGAGCCCACACUGCCGCAGCCACAGCCUCGCAGC-CGCUUGG----------- .((((((((((((((.((((((((....)))))))).).....)))))..))))------.)).))(((..(.((.((.((....))...)).))-.)..)))----------- ( -30.70, z-score = -0.64, R) >droGri2.scaffold_15245 18050480 90 + 18325388 CGCCCUUUCUGUAUAUGUUGCAACAGAUGCUACAGCCUAACUUGAAUGUGGACA------UGGUAG--ACUCCUCGUCAG-UGCAAC---GUAAU-CGAAUGG----------- ...((.(((.((.(((((((((((..((((((((..(......)..))))).))------).)).(--((.....)))..-))))))---)))))-.))).))----------- ( -23.50, z-score = -1.10, R) >consensus CACCGAUGCUCUACAUGCUGCAACAGCUGUUGCAGCCCAACUUUAAGGUGAAUAUCU___UGGUGGAACCACCAAAGCAGCCACAGCGUCGCAUUA__________________ ................((((((((....)))))))).............................................................................. ( -9.45 = -9.65 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:43 2011