
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,947,485 – 14,947,608 |
| Length | 123 |
| Max. P | 0.808831 |

| Location | 14,947,485 – 14,947,585 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 59.98 |
| Shannon entropy | 0.73623 |
| G+C content | 0.52586 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -11.28 |
| Energy contribution | -11.94 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14947485 100 + 21146708 ------GCUCCACA-ACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUUCAGCUAGUGGAGGAUCUGCU------------ ------.((((((.-....((((.(((.((...........)).))).))))..(((.(((((........)))))..)))............))))))........------------ ( -27.60, z-score = -1.28, R) >droSim1.chr2R 13647028 100 + 19596830 ------GCUCCACA-ACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUAGUGGAGGAUCUGCU------------ ------.((((((.-.........(((..............))).(((..(((((((..............)))))))(((....))))))..))))))........------------ ( -29.08, z-score = -1.71, R) >droSec1.super_1 12453988 97 + 14215200 ------GCUACACA-ACCACCACCGCAUUAUCAUC---ACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCUGGAUGCCGGAAAAUUCCAGCUAGUGGAGGAUCUGCU------------ ------........-.((.((((.(((........---...))).(((..((((((((............))))))))(((....))))))..)))).)).......------------ ( -27.90, z-score = -1.47, R) >droYak2.chr2R 6903172 100 - 21139217 ------GCUCCACA-ACCACCACCGCAUUAUCAUCCUCACUUGCAGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUGGUCGAGGAUCUAAU------------ ------........-.................((((((((...(((((..(((((((..............)))))))(((....)))))))))).)))))).....------------ ( -28.44, z-score = -2.22, R) >droEre2.scaffold_4845 9144651 100 + 22589142 ------GCUCCACG-ACCACCACCACAUUAUCAUCCUCACUUGCUGCUGUGGCAUCCACAUCCCCGCCGCCGGAUGCCGGAAAAUUCCAGCUGGUGGAGGAUCUAAU------------ ------.......(-(((.((((((......((........))..(((..(((((((..............)))))))(((....)))))))))))).)).))....------------ ( -28.74, z-score = -1.14, R) >droWil1.scaffold_180697 2593263 117 + 4168966 --GAAGACUGCAAAUGGAACCACCACUGAGAAGGCUGAAUUAAUUCCAUCUGUUACUGCUUCAUCGAUGCCAUUUGCUGGGGCUUUUGGAAAGGUGGAGCUGAUUUUGGAUGAUGGACU --.......((...(((.....)))((....)))).........((((((.(((...((((((((....(((...((....))...)))...))))))))........))))))))).. ( -26.90, z-score = 0.50, R) >droGri2.scaffold_15245 18050029 83 + 18325388 GUCAAGACUACGGACAAAACUG---------AAACUGACGUUGCCACAGCCACAAAUGCUGCAGCAGAUGCAGUUGCUGAGA------AAAAGCUGCA--------------------- ((((......(((......)))---------....)))).((((..((((.......))))..)))).(((((((.......------...)))))))--------------------- ( -19.00, z-score = 0.40, R) >consensus ______GCUCCACA_ACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUGGUGGAGGAUCUGCU____________ ................((.((((...........................(((((((..............)))))))(((....))).....)))).))................... (-11.28 = -11.94 + 0.66)
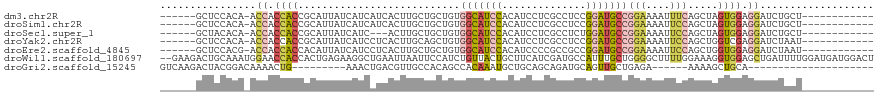
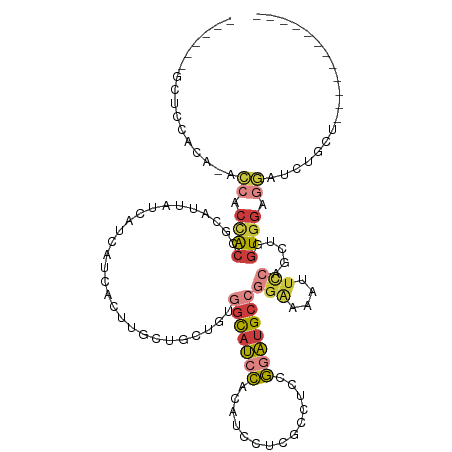

| Location | 14,947,485 – 14,947,585 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 59.98 |
| Shannon entropy | 0.73623 |
| G+C content | 0.52586 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -14.66 |
| Energy contribution | -16.05 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14947485 100 - 21146708 ------------AGCAGAUCCUCCACUAGCUGAAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGAUGAUAAUGCGGUGGUGGU-UGUGGAGC------ ------------........((((((.((((.......((((.(((((........))))))))).(((((.(((.((........))....))).))))))))-))))))).------ ( -34.30, z-score = -1.09, R) >droSim1.chr2R 13647028 100 - 19596830 ------------AGCAGAUCCUCCACUAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGAUGAUAAUGCGGUGGUGGU-UGUGGAGC------ ------------........((((((.((((.......((((.(((((........))))))))).(((((.(((.((........))....))).))))))))-))))))).------ ( -34.30, z-score = -0.85, R) >droSec1.super_1 12453988 97 - 14215200 ------------AGCAGAUCCUCCACUAGCUGGAAUUUUCCGGCAUCCAGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGU---GAUGAUAAUGCGGUGGUGGU-UGUGUAGC------ ------------.(((.(.((.(((((.((((((....)))((((((((.(........).))))))))..))).(((...---........)))))))).)).-).)))...------ ( -35.80, z-score = -1.82, R) >droYak2.chr2R 6903172 100 + 21139217 ------------AUUAGAUCCUCGACCAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCUGCAAGUGAGGAUGAUAAUGCGGUGGUGGU-UGUGGAGC------ ------------.(((.(((((((.((.((((((....))))))....))...))))))).)))...(((((((..(......(.((....)).)....)..))-)))))...------ ( -37.00, z-score = -1.93, R) >droEre2.scaffold_4845 9144651 100 - 22589142 ------------AUUAGAUCCUCCACCAGCUGGAAUUUUCCGGCAUCCGGCGGCGGGGAUGUGGAUGCCACAGCAGCAAGUGAGGAUGAUAAUGUGGUGGUGGU-CGUGGAGC------ ------------....((.((.((((((((((((....))))))((((.((.((...(.(((((...))))).).))..))..)))).......)))))).)))-).......------ ( -36.70, z-score = -0.98, R) >droWil1.scaffold_180697 2593263 117 - 4168966 AGUCCAUCAUCCAAAAUCAGCUCCACCUUUCCAAAAGCCCCAGCAAAUGGCAUCGAUGAAGCAGUAACAGAUGGAAUUAAUUCAGCCUUCUCAGUGGUGGUUCCAUUUGCAGUCUUC-- ......(((((........(((.............))).(((.....)))....)))))........(((((((((((......(((........))))))))))))))........-- ( -20.02, z-score = 0.76, R) >droGri2.scaffold_15245 18050029 83 - 18325388 ---------------------UGCAGCUUUU------UCUCAGCAACUGCAUCUGCUGCAGCAUUUGUGGCUGUGGCAACGUCAGUUU---------CAGUUUUGUCCGUAGUCUUGAC ---------------------(((((.....------.........)))))((.(((((.(((.(((..((((((....)).))))..---------)))...)))..)))))...)). ( -20.24, z-score = 0.84, R) >consensus ____________AGCAGAUCCUCCACCAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAAGAUGAUAAUGCGGUGGUGGU_UGUGGAGC______ ....................((((((..............(.((((((........)))))).)..(((((.((.(((..............))).)).)))))..))))))....... (-14.66 = -16.05 + 1.40)
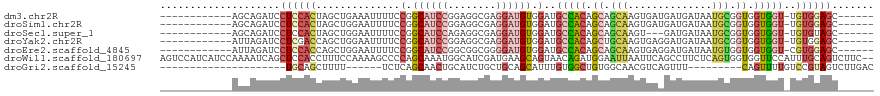
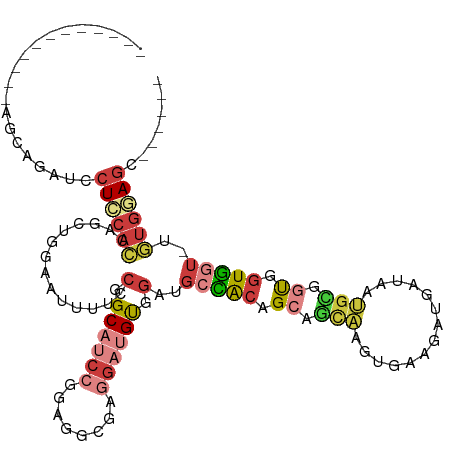
| Location | 14,947,511 – 14,947,608 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.65 |
| Shannon entropy | 0.67013 |
| G+C content | 0.56121 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14947511 97 + 21146708 -----UCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUUCAGCUAGU---GGAGGAUCUGCUGGGACUGCGCCGUCCCAAGAGGC--------------- -----((..((((.((((.(.((((((((..............)))))))).)......)))).)))---)..)).(((..((((((......))))))..))).--------------- ( -33.04, z-score = -0.22, R) >droSim1.chr2R 13647054 97 + 19596830 -----UCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUAGU---GGAGGAUCUGCUGGGACUGCGCCGUCCCAAGAGGC--------------- -----..............(((((...))))).((((.((((((...((.(((....))).))...)---)))))......((((((......)))))).)))).--------------- ( -34.70, z-score = -0.60, R) >droSec1.super_1 12454011 97 + 14215200 -----UCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCUGGAUGCCGGAAAAUUCCAGCUAGU---GGAGGAUCUGCUGGGACUGCGCCGUCCCAAGAGGC--------------- -----((..((((.(((.....((((((((............))))))))(((....)))))).)))---)..)).(((..((((((......))))))..))).--------------- ( -35.50, z-score = -0.87, R) >droYak2.chr2R 6903198 97 - 21139217 -----UCCUCACUUGCAGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUGGU---CGAGGAUCUAAUGGGACUGCGCCGUCCCAAGAGGC--------------- -----.((((.(((((((((..(((((((..............)))))))(((....))))))))..---)))).......((((((......)))))).)))).--------------- ( -39.24, z-score = -2.30, R) >droEre2.scaffold_4845 9144677 97 + 22589142 -----UCCUCACUUGCUGCUGUGGCAUCCACAUCCCCGCCGCCGGAUGCCGGAAAAUUCCAGCUGGU---GGAGGAUCUAAUGGGACUGCGACGUCCCAAGAGGC--------------- -----((((((((.(((.....(((((((..............)))))))(((....)))))).)))---.)))))(((..((((((......))))))..))).--------------- ( -39.04, z-score = -1.84, R) >droWil1.scaffold_180697 2593303 115 + 4168966 -----AUUCCAUCUGUUACUGCUUCAUCGAUGCCAUUUGCUGGGGCUUUUGGAAAGGUGGAGCUGAUUUUGGAUGAUGGACUACCAAUGGAACUAACCAAAUUGCAUCAUGGCAAACGUC -----..((((...((((..((((((((....(((...((....))...)))...))))))))))))..)))).((((.........(((......)))..((((......)))).)))) ( -29.80, z-score = -0.49, R) >droGri2.scaffold_15245 18050048 89 + 18325388 CUGAAACUGACGUUGCCACAGCCACAAAUGCUGCAGCAGAUGCAGUUGCUGAGAAAAAGCUGCAGCUGCUGGAUGAUUUUCAGCCCAGU------------------------------- ((((((....((((....((((.......))))((((((.(((((((..........))))))).))))))))))..))))))......------------------------------- ( -28.50, z-score = -0.66, R) >consensus _____UCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCUGGU___GGAGGAUCUACUGGGACUGCGCCGUCCCAAGAGGC_______________ ..........(((.((((.((((.....))))........(((((...)))))......)))).))).........(((..((((((......))))))..)))................ (-14.27 = -14.63 + 0.36)


| Location | 14,947,511 – 14,947,608 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.65 |
| Shannon entropy | 0.67013 |
| G+C content | 0.56121 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -16.47 |
| Energy contribution | -18.13 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14947511 97 - 21146708 ---------------GCCUCUUGGGACGGCGCAGUCCCAGCAGAUCCUCC---ACUAGCUGAAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGA----- ---------------..((.(((((((......))))))).))......(---(((.((((.........((((((((.(........).))))))))....)))).))))....----- ( -35.52, z-score = -0.71, R) >droSim1.chr2R 13647054 97 - 19596830 ---------------GCCUCUUGGGACGGCGCAGUCCCAGCAGAUCCUCC---ACUAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGA----- ---------------.(((((((((((......))))))).....(((((---....((((((....))))))....))))).)))).(((((...)))))..............----- ( -38.00, z-score = -1.06, R) >droSec1.super_1 12454011 97 - 14215200 ---------------GCCUCUUGGGACGGCGCAGUCCCAGCAGAUCCUCC---ACUAGCUGGAAUUUUCCGGCAUCCAGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGA----- ---------------..((.(((((((......))))))).))......(---(((.((((((....)))((((((((.(........).)))))))).....))).))))....----- ( -38.30, z-score = -1.44, R) >droYak2.chr2R 6903198 97 + 21139217 ---------------GCCUCUUGGGACGGCGCAGUCCCAUUAGAUCCUCG---ACCAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCUGCAAGUGAGGA----- ---------------..((..((((((......))))))..)).((((((---..((((((((....)))((((((((.(........).))))))))..)))))....))))))----- ( -43.60, z-score = -2.60, R) >droEre2.scaffold_4845 9144677 97 - 22589142 ---------------GCCUCUUGGGACGUCGCAGUCCCAUUAGAUCCUCC---ACCAGCUGGAAUUUUCCGGCAUCCGGCGGCGGGGAUGUGGAUGCCACAGCAGCAAGUGAGGA----- ---------------..((..((((((......))))))..)).(((((.---((..((((((....)))((((((((.((.......)))))))))).....)))..)))))))----- ( -39.10, z-score = -1.06, R) >droWil1.scaffold_180697 2593303 115 - 4168966 GACGUUUGCCAUGAUGCAAUUUGGUUAGUUCCAUUGGUAGUCCAUCAUCCAAAAUCAGCUCCACCUUUCCAAAAGCCCCAGCAAAUGGCAUCGAUGAAGCAGUAACAGAUGGAAU----- (((..((((......))))....))).((((((((..((.(.(.(((((........(((.............))).(((.....)))....))))).).).))...))))))))----- ( -18.82, z-score = 1.73, R) >droGri2.scaffold_15245 18050048 89 - 18325388 -------------------------------ACUGGGCUGAAAAUCAUCCAGCAGCUGCAGCUUUUUCUCAGCAACUGCAUCUGCUGCAGCAUUUGUGGCUGUGGCAACGUCAGUUUCAG -------------------------------.(((((.((.....)))))))..((((((((.......(((...))).....))))))))...((..((((((....)).))))..)). ( -30.70, z-score = -0.98, R) >consensus _______________GCCUCUUGGGACGGCGCAGUCCCAGCAGAUCCUCC___ACCAGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGA_____ .....................((((((......))))))..................((((......(((.((((((........))))))))).....))))................. (-16.47 = -18.13 + 1.66)

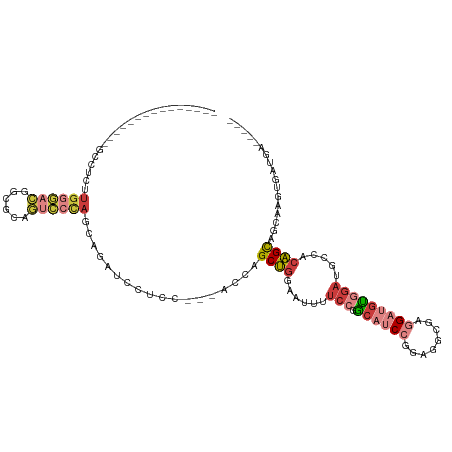
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:43 2011