| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,929,926 – 14,930,026 |
| Length | 100 |
| Max. P | 0.787111 |

| Location | 14,929,926 – 14,930,026 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.45 |
| Shannon entropy | 0.50343 |
| G+C content | 0.51191 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.86 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14929926 100 - 21146708 ---GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUCCUGUGAAAU-CCCCUCAGCUUCCU-UCCCUUCCCAUCACAUCCUUUUCU-- ---(((((((.(..((((((.((((((.(.........))))))..(((((.......)))))..-..).))))))..).-...........)))))))......-- ( -22.82, z-score = -1.64, R) >droSim1.chr2R 13629457 102 - 19596830 ---GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAACUGGCACUUCAUCUAUCCUGUGAAAU-CCCCUCAGCUUCCU-GCCCAUCCUUUCCCUUCCUCUUCUUC ---(((((.(((..((((((.((((((............)))))..(((((.......)))))..-..).))))))..).-)).))))).................. ( -25.50, z-score = -3.08, R) >droSec1.super_1 12436525 102 - 14215200 ---GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUCCUGUGAAAU-CCCUUCAGCUUCCU-GCCCAUCCCUUCCCUUCCUCUUCUUC ---(((((.(((..((((((.((((((.(.........))))))..(((((.......)))))..-..).))))))..).-)).))))).................. ( -25.40, z-score = -2.30, R) >droYak2.chr2R 6885039 95 + 21139217 ---GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUCCUGUGAAAU-CCCCUCAGCUUCCUAACCCUUCCUUUUAUCUCC-------- ---(((.((..(..((((((.((((((.(.........))))))..(((((.......)))))..-..).))))))..)..))...)))..........-------- ( -18.60, z-score = -0.42, R) >droEre2.scaffold_4845 9126693 93 - 22589142 ---GGAAGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUCCUGUGAAAU-CCCCCCAGCUUCUU---CCUUCCCCUCCUCUUCU------- ---(((((.((((((((((..(((.((....)))))))))))))..(((((.......)))))..-.......))..)))---))...............------- ( -20.90, z-score = -1.01, R) >droAna3.scaffold_13266 2497409 82 + 19884421 UGGAGAAGUGCGUGAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUUUAUCCUGUGCAAUAUCCCAUUGCUUUUCCU------------------------- .((((((((((...(((((..(((.((....)))))))))).)))))))...........(((((.....)))))...))).------------------------- ( -22.00, z-score = -0.73, R) >droPer1.super_2 6203366 97 - 9036312 ---GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUUUUGUACCCCCCAUUGCCACGCCCU-GCUGGUGCUGAUGCUGCUGC------ ---((..(((((.(((.(((..(((((.(.........))))))...))).)))...)))))..))((..((((.((...-))))))..))..........------ ( -24.20, z-score = 1.14, R) >consensus ___GGAUGUGCGUUAGUUGAUGGCCAGACACCUGCUUAGCUGGCACUUCAUCUAUCCUGUGAAAU_CCCCUCAGCUUCCU_GCCCUUCCCUUCCCUUCCU_______ ...(((((.(.((((((((..(((.((....))))))))))))).)..)))))...................................................... (-13.79 = -13.86 + 0.06)

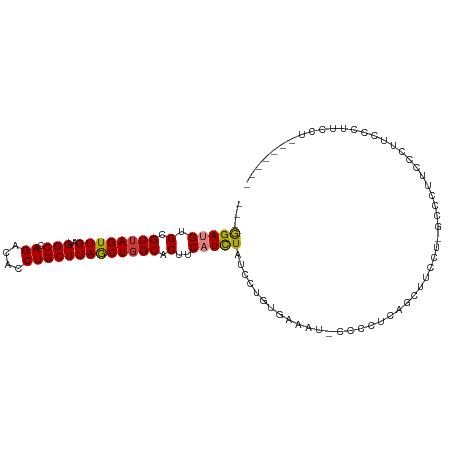
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:38 2011