| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,245,870 – 3,246,006 |
| Length | 136 |
| Max. P | 0.734338 |

| Location | 3,245,870 – 3,246,006 |
|---|---|
| Length | 136 |
| Sequences | 3 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 62.56 |
| Shannon entropy | 0.50711 |
| G+C content | 0.48807 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
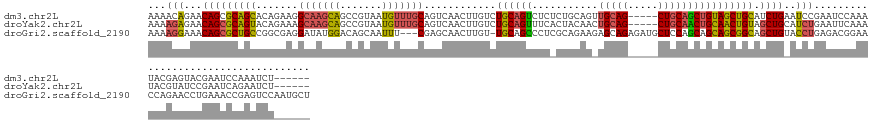
>dm3.chr2L 3245870 136 + 23011544 AAAACAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUCUCUCUGCAGUUGCAG-----CUGCAGCUGUAGCUGCAUCUGAAUCCGAAUCCAAAUACGAGUACGAAUCCAAAUCU------ ....((((.....((((.(((((..(((((((.......)))))))..))....)))))))........(((((((((((-----(....))))))))))))))))...................................------ ( -42.20, z-score = -1.64, R) >droYak2.chr2L 3235695 136 + 22324452 AAAAGAGAACAGCGCAGUACAGAAAGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUUUCACUACAACUGCAG-----CUGCAACUGCAACUGUAGCUGCAUCUGAAUUCAAAUACGUAUCCGAAUCAGAAUCU------ .....(((.(((((((((.(((...(((((((.......))))))).......((((((((((((.......))))))))-----..)))))))..))))).))))..(((((.(((............)))))))).)))------ ( -38.80, z-score = -1.54, R) >droGri2.scaffold_2190 1013 143 - 1372 AAAAGGAAACAGCGCUGCCGGCGAGGAUAUGGACAGCAAUUU---CGAGCAACUUGU-UGCAGCCCUCGCAGAAGAGCAGAGAUGCUCCAGCAGCAGCGGCAGCUGUACCUGAGACGGAACCAGAACCUGAAACCGAGUCCAAUGCU ....(....)((((((((((((((((.....((.(....).)---)..((((....)-)))...))))))....(((((....))))).........))))))).......((..(((...(((...)))...)))..))....))) ( -45.40, z-score = -0.72, R) >consensus AAAAGAGAACAGCGCAGCACAGAAGGCAAGCAGCCGUAAUGUUUGCAGUCAACUUGUCUGCAGUCUCACUACAACUGCAG_____CUGCAGCUGCAGCUGCAGCUGAAUCUGAAUCCAAAUACGAAUCCGAAUCCGAAUCU______ ...(((...(((((((((.......(((((((.......)))))))............((((((...........(((((.....)))))))))))))))).))))..))).................................... (-22.81 = -23.27 + 0.46)
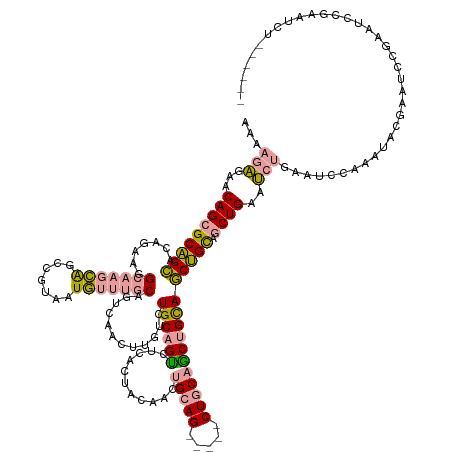

| Location | 3,245,870 – 3,246,006 |
|---|---|
| Length | 136 |
| Sequences | 3 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 62.56 |
| Shannon entropy | 0.50711 |
| G+C content | 0.48807 |
| Mean single sequence MFE | -48.80 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
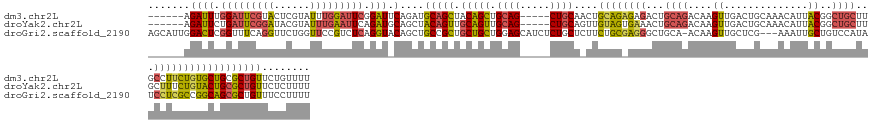
>dm3.chr2L 3245870 136 - 23011544 ------AGAUUUGGAUUCGUACUCGUAUUUGGAUUCGGAUUCAGAUGCAGCUACAGCUGCAG-----CUGCAACUGCAGAGAGACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUGUUUU ------..(((((((((((.(..((....))..).)))))))))))((((..(((((.((((-----(.(((.((((((.....)))))).(((((.(.(((.........)))).))))).....))))))))))))).))))... ( -47.40, z-score = -1.52, R) >droYak2.chr2L 3235695 136 - 22324452 ------AGAUUCUGAUUCGGAUACGUAUUUGAAUUCAGAUGCAGCUACAGUUGCAGUUGCAG-----CUGCAGUUGUAGUGAAACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCUUUCUGUACUGCGCUGUUCUCUUUU ------....((((((((((((....))))))).))))).(((((..((((.((((..((((-----..((((((((((((....(((((.(.....).)))))..))))))))))))))))...)))))))).)))))........ ( -49.70, z-score = -2.88, R) >droGri2.scaffold_2190 1013 143 + 1372 AGCAUUGGACUCGGUUUCAGGUUCUGGUUCCGUCUCAGGUACAGCUGCCGCUGCUGCUGGAGCAUCUCUGCUCUUCUGCGAGGGCUGCA-ACAAGUUGCUCG---AAAUUGCUGUCCAUAUCCUCGCCGGCAGCGCUGUUUCCUUUU ......(((..((((....(((.(((...((......))..)))..)))(((((((..((((((....))))))...(((((((..(((-(....))))..(---(........))....))))))))))))))))))..))).... ( -49.30, z-score = -0.46, R) >consensus ______AGAUUCGGAUUCGGAUUCGUAUUUGGAUUCAGAUACAGCUGCAGCUGCAGCUGCAG_____CUGCAAUUGUAGAGAGACUGCAGACAAGUUGACUGCAAACAUUACGGCUGCUUGCCUUCUGUGCUGCGCUGUUCUCUUUU .......((((.(((((((((......))))))))).))).)....(((((.(((((.((((.....))))....((((((((...((((....((..............))..))))...))))))))))))))))))........ (-22.74 = -23.87 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:24 2011