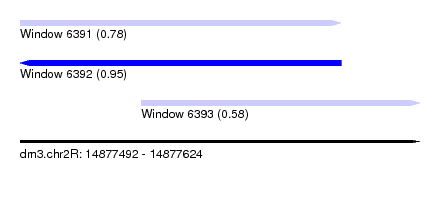
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,877,492 – 14,877,624 |
| Length | 132 |
| Max. P | 0.945844 |

| Location | 14,877,492 – 14,877,598 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.36467 |
| G+C content | 0.40972 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -18.87 |
| Energy contribution | -19.98 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
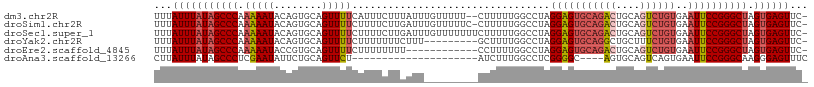
>dm3.chr2R 14877492 106 + 21146708 UUUAUUUAUAGCCCAAAAAUACAGUGCAGUUUUCAUUUCUUUAUUUGUUUUU--CUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUUC- ...(((((((((((.....(((((((((((((.(((((((............--...........))))))).))))))))..)))))......))))).))))))..- ( -30.20, z-score = -2.99, R) >droSim1.chr2R 13576425 107 + 19596830 UUUAUUUAUAGCCCAAAAAUACAGUGCAGUUUUCUUUUCUUGAUUUGUUUUUC-CUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUUC- ...(((((((((((.....((((((((((((.((.......))..(((..(((-((.........))))).))))))))))..)))))......))))).))))))..- ( -27.60, z-score = -1.58, R) >droSec1.super_1 12383704 108 + 14215200 UUUAUUUAUAGCCCAAAAAUACAGUGCAGUUUUCUUUUCUUGAUUUGUUUUUUUCUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUUC- ...((((((((((((((((.((((..(((..........)))..)))).)))))............(((((((((((....))))))..)))))))))).))))))..- ( -27.30, z-score = -1.59, R) >droYak2.chr2R 6822553 99 - 21139217 UUUAUUUAUAGCCCAAAAAUACAGUGCAGUUUUCUUUUUUUCUUU---------GCUUUUGGCCUAGGAGUGCAGGCUGCUUUCUGUGAAUUCCGGGCUAGUGAGUUC- ...(((((((((((.....(((((.(((((((.(....(((((..---------((.....))..))))).).)))))))...)))))......))))).))))))..- ( -28.50, z-score = -1.81, R) >droEre2.scaffold_4845 9073099 96 + 22589142 UUUAUUUAUAGCCCAAAAAUACCGUGCAGUUUUCUUUUUUUU------------CCUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUUC- ...(((((((((((............................------------((....))....(((((((((((....))))))..)))))))))).))))))..- ( -25.60, z-score = -1.80, R) >droAna3.scaffold_13266 2425972 84 - 19884421 CUUAUUUAUAGCCCUCGAAUAUUCUGCAGUUCU---------------------AUCUUUGGCCUCGGGGC----AGUGCAGUCAGUGAAUUCCGGGCAAGGGAGUUUC ...........((((.((((........)))).---------------------.......(((.((((.(----(.((....)).))...))))))).))))...... ( -18.70, z-score = 0.92, R) >consensus UUUAUUUAUAGCCCAAAAAUACAGUGCAGUUUUCUUUUCUUGAUU_________CUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUUC_ ...(((((((((((.(((((........))))).................................(((((((((((....))))))..)))))))))).))))))... (-18.87 = -19.98 + 1.11)
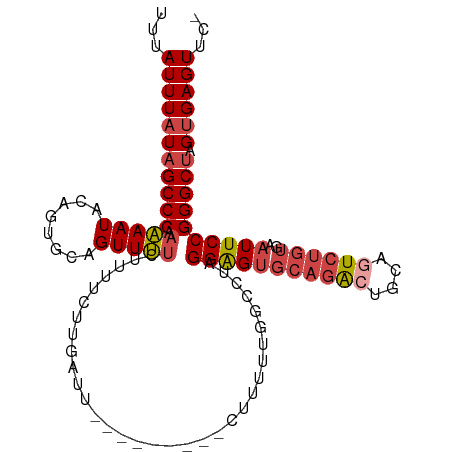
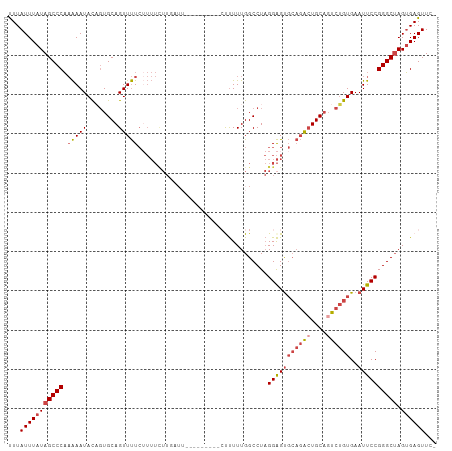
| Location | 14,877,492 – 14,877,598 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.36467 |
| G+C content | 0.40972 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -15.73 |
| Energy contribution | -17.18 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14877492 106 - 21146708 -GAACUCACUAGCCCGGAAUUCACAGACUGCAGUCUGCACUCCUAGGCCAAAAAG--AAAAACAAAUAAAGAAAUGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAA -........((((((((((...((((..((((((..((........)).......--............(....)....))))))))))..))))))))))........ ( -23.00, z-score = -2.35, R) >droSim1.chr2R 13576425 107 - 19596830 -GAACUCACUAGCCCGGAAUUCACAGACUGCAGUCUGCACUCCUAGGCCAAAAAG-GAAAAACAAAUCAAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAA -........((((((((((...((((..((((((......((((.........))-))........((.......))..))))))))))..))))))))))........ ( -25.90, z-score = -2.66, R) >droSec1.super_1 12383704 108 - 14215200 -GAACUCACUAGCCCGGAAUUCACAGACUGCAGUCUGCACUCCUAGGCCAAAAAGAAAAAAACAAAUCAAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAA -........((((((((((...((((..((((((..((........))..................((.......))..))))))))))..))))))))))........ ( -23.60, z-score = -2.41, R) >droYak2.chr2R 6822553 99 + 21139217 -GAACUCACUAGCCCGGAAUUCACAGAAAGCAGCCUGCACUCCUAGGCCAAAAGC---------AAAGAAAAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAA -........((((((((((...((((......(((((......))))).....((---------(................))).))))..))))))))))........ ( -24.59, z-score = -2.32, R) >droEre2.scaffold_4845 9073099 96 - 22589142 -GAACUCACUAGCCCGGAAUUCACAGACUGCAGUCUGCACUCCUAGGCCAAAAGG------------AAAAAAAAGAAAACUGCACGGUAUUUUUGGGCUAUAAAUAAA -........(((((((((((((.(((((....)))))...((((........)))------------).......))).(((....)))..))))))))))........ ( -23.90, z-score = -2.11, R) >droAna3.scaffold_13266 2425972 84 + 19884421 GAAACUCCCUUGCCCGGAAUUCACUGACUGCACU----GCCCCGAGGCCAAAGAU---------------------AGAACUGCAGAAUAUUCGAGGGCUAUAAAUAAG ...........((((.((((.......((((...----(((....)))...((..---------------------....))))))...))))..)))).......... ( -16.20, z-score = -0.32, R) >consensus _GAACUCACUAGCCCGGAAUUCACAGACUGCAGUCUGCACUCCUAGGCCAAAAAG_________AAUAAAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAA .........((((((((((...((((..((((((.............................................))))))))))..))))))))))........ (-15.73 = -17.18 + 1.45)
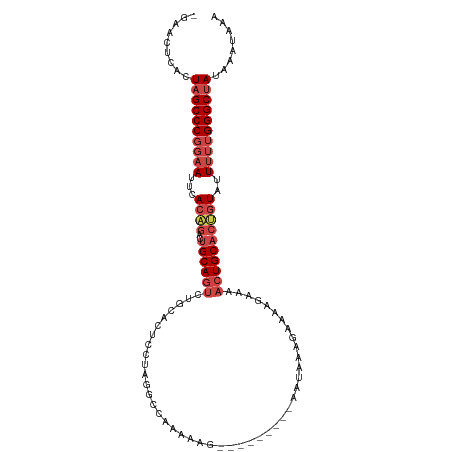
| Location | 14,877,532 – 14,877,624 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Shannon entropy | 0.34394 |
| G+C content | 0.46156 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -19.58 |
| Energy contribution | -20.67 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14877532 92 + 21146708 UUAUUUGUUUUUC--UUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUU-CUAUUUUCGGCAU-AUUCCCAUAGGGAG ............(--((.(((((((.(((((((((((....))))))..)))))))))))).)))..-.............-..((((...)))). ( -27.60, z-score = -1.87, R) >droSim1.chr2R 13576465 93 + 19596830 UGAUUUGUUUUUC-CUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUU-CUAUUUUCGGCAU-AUUCCCAUAGGGAG .............-(((.(((((((.(((((((((((....))))))..)))))))))))).)))..-.............-..((((...)))). ( -27.80, z-score = -1.42, R) >droSec1.super_1 12383744 94 + 14215200 UGAUUUGUUUUUUUCUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUU-CUAUUUUCGGCAU-AUUCCCAUAGGGAG ..............(((.(((((((.(((((((((((....))))))..)))))))))))).)))..-.............-..((((...)))). ( -27.60, z-score = -1.57, R) >droYak2.chr2R 6822593 85 - 21139217 ---------UCUUUGCUUUUGGCCUAGGAGUGCAGGCUGCUUUCUGUGAAUUCCGGGCUAGUGAGUU-CUAUUUUCGGUAU-AUUCCCAAAGGGAG ---------.....(((((((((((.((((((((((......)))))..)))))))))))).)))).-.............-..((((...)))). ( -23.20, z-score = 0.01, R) >droEre2.scaffold_4845 9073138 83 + 22589142 -----------UUUCCUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUU-CUAUUUUCGGUAU-AUUCCCAUAGGGAG -----------..(((((.((((((.(((((((((((....))))))..)))))))))((.((((..-.....)))).)).-.....)).))))). ( -26.90, z-score = -1.77, R) >droAna3.scaffold_13266 2426002 79 - 19884421 -----------UCUAUCUUUGGCCUCGGGG--CAG--UGCAGUCAGUGAAUUCCGGGCAAGGGAGUUUCUAUUUUCAAUCCCACUCCCAUAAGG-- -----------.....((((((((.((((.--((.--((....)).))...)))))))..((((((................)))))).)))))-- ( -21.29, z-score = -0.41, R) >consensus _________UUUCUCUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGUU_CUAUUUUCGGCAU_AUUCCCAUAGGGAG ..................(((((((.(((((((((((....))))))..)))))))))))).(((........)))........(((.....))). (-19.58 = -20.67 + 1.08)

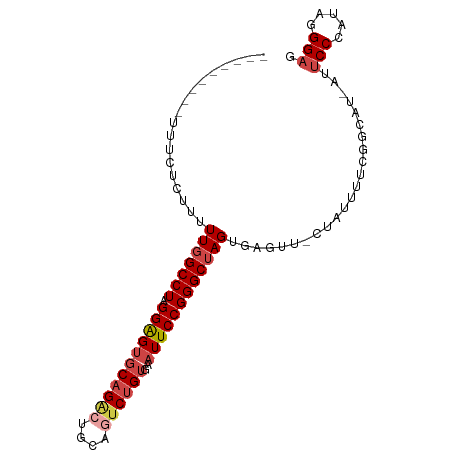
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:30 2011