
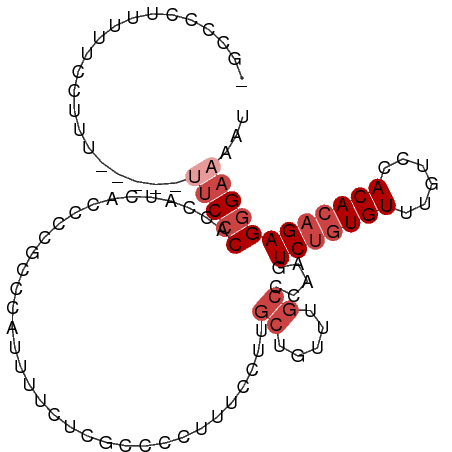
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,871,316 – 14,871,438 |
| Length | 122 |
| Max. P | 0.785887 |

| Location | 14,871,316 – 14,871,414 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.38615 |
| G+C content | 0.52006 |
| Mean single sequence MFE | -17.19 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.93 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14871316 98 + 21146708 -GCCGCUUUUUCCCUU----UUUCCCACCAUCACUCCCACCAUUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUGCACACAGAGGGAAAAU -..............(----((((((...................................((.....)).....(((((((......)))))))))))))). ( -18.20, z-score = -1.68, R) >droSim1.chr2R 13570250 97 + 19596830 -GCCCCUUUUUCCUUU-----UUCCCACCAUCACCCCGCCCAUUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAU -.............((-----(((((...................................((.....)).....(((((((......)))))))))))))). ( -18.20, z-score = -2.67, R) >droSec1.super_1 12377553 97 + 14215200 -GCCCCUUUUUCCUUU-----UUCCCACCAUCACCCCGCCCACUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAU -.............((-----(((((...................................((.....)).....(((((((......)))))))))))))). ( -18.20, z-score = -2.50, R) >droYak2.chr2R 6816187 102 - 21139217 -GCCCCUUUUUCCUUUCCUUUUUCCCACCAUCACCCCGCCCAUUUUUCCGCCCCUUUCCUUGCUGUUUGCCAAAGUCUAUGUUUGUCCACACAGAGGGAAAUC -.....................................................(((((((.((((.((.((((.......))))..)).))))))))))).. ( -12.10, z-score = -0.63, R) >droEre2.scaffold_4845 9066743 92 + 22589142 -GCCCCUUUUUCCUUU------UCCCACCAACACCCCGCCCAUUUUUCCGCCCCUUUCCUUGCUGUUUGCCAAAGUCUGUGUUUGUCCACACAGAGGGA---- -...............------((((...................................((.....)).....(((((((......)))))))))))---- ( -14.90, z-score = -1.25, R) >droAna3.scaffold_13266 2419727 86 - 19884421 GGCUCCUUGGCCCUUU------ACCCGGCCUCAACCC-----------CACCUCCUCGUUCAGUGUUUGCCUUAGUCUGUGUUUGCCGACACUGAGAGAAAAU ((......((((....------....))))......)-----------).....(((..((((((((.((..............)).)))))))))))..... ( -21.54, z-score = -1.50, R) >consensus _GCCCCUUUUUCCUUU_____UUCCCACCAUCACCCCGCCCAUUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAU .....................(((((...................................((.....)).....(((((((......))))))))))))... (-10.73 = -11.93 + 1.21)

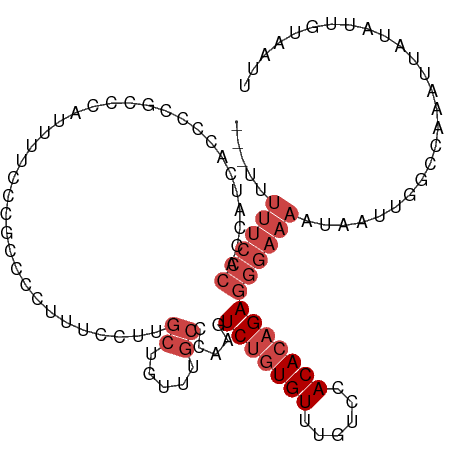
| Location | 14,871,329 – 14,871,438 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.49724 |
| G+C content | 0.44163 |
| Mean single sequence MFE | -17.79 |
| Consensus MFE | -10.93 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
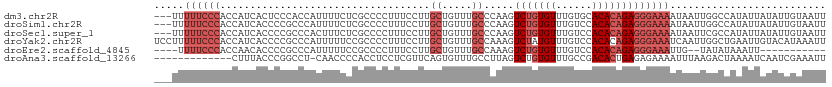
>dm3.chr2R 14871329 109 + 21146708 ---UUUUUCCCACCAUCACUCCCACCAUUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUGCACACAGAGGGAAAAUAAUUGGCCAUAUUAUAUUGUAAUU ---...............................(((..(((((((.((((.(((.((((......)))).))).)))))))))))......)))................. ( -19.90, z-score = -1.02, R) >droSim1.chr2R 13570262 109 + 19596830 ---UUUUUCCCACCAUCACCCCGCCCAUUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAUAAUUGGCCAUAUUAUAUUGUAAUU ---...................(((.((((((((............((.....)).....(((((((......)))))))))))))))....)))................. ( -19.80, z-score = -1.39, R) >droSec1.super_1 12377565 109 + 14215200 ---UUUUUCCCACCAUCACCCCGCCCACUUUCUCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAUAAUUCGCCAUAUUAUAUUGUAAUU ---.(((((((...................................((.....)).....(((((((......))))))))))))))......................... ( -18.20, z-score = -1.80, R) >droYak2.chr2R 6816201 112 - 21139217 UCCUUUUUCCCACCAUCACCCCGCCCAUUUUUCCGCCCCUUUCCUUGCUGUUUGCCAAAGUCUAUGUUUGUCCACACAGAGGGAAAUCAAUUGGCUGAAUUGUACAUAAAUU ......................(..((...(((.(((..(((((((.((((.((.((((.......))))..)).)))))))))))......))).))).))..)....... ( -17.90, z-score = -0.37, R) >droEre2.scaffold_4845 9066755 95 + 22589142 ----UUUUCCCACCAACACCCCGCCCAUUUUUCCGCCCCUUUCCUUGCUGUUUGCCAAAGUCUGUGUUUGUCCACACAGAGGGAAAUUG--UAUAUAAAUU----------- ----.((((((...................................((.....)).....(((((((......)))))))))))))...--..........----------- ( -17.30, z-score = -1.82, R) >droAna3.scaffold_13266 2419739 98 - 19884421 -------------CUUUACCCGGCCU-CAACCCCACCUCCUCGUUCAGUGUUUGCCUUAGUCUGUGUUUGCCGACACUGAGAGAAAAUUUAAGACUAAAAUCAAUCGAAAUU -------------........((...-.....)).....(((..((((((((.((..............)).)))))))))))............................. ( -13.64, z-score = 0.29, R) >consensus ___UUUUUCCCACCAUCACCCCGCCCAUUUUCCCGCCCCUUUCCUUGCUGUUUGCCCAAGUCUGUGUUUGUCCACACAGAGGGAAAAUAAUUGGCCAAAUUAUAUUGUAAUU .....((((((...................................((.....)).....(((((((......))))))))))))).......................... (-10.93 = -12.60 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:27 2011