
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,862,386 – 14,862,496 |
| Length | 110 |
| Max. P | 0.806192 |

| Location | 14,862,386 – 14,862,496 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.59511 |
| G+C content | 0.62124 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806192 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 14862386 110 + 21146708 CCAGCGGCGGUUGCCAGCGAG-GAACCAGAGGUCAUCGAGAUC---GACGACGGCGAUGCGGCCGCAAAUGCAGAGGUGGCGGCCAUCAAUGAUGGCAGCAACACCAGUAGGAG----- ((.(((((.((((((.....(-....)....(((.(((....)---)).)))))))..)).))))).........((((((.((((((...)))))).))..))))....))..----- ( -43.70, z-score = -2.40, R) >droSim1.chr2R 13561081 110 + 19596830 CCAGCGGCGGUUGCCAGCGAG-GAAGCGGAAGUCAUCGAGAUC---GACGACGGCGAUGCGGCCGCAAAUGCAGAGGUGGCGGCCAUCAACGAUGGCAGCAACACCAGCAGGAG----- ((.(((((.((((((.((...-...))....(((.(((....)---)).)))))))..)).)))))...(((...((((((.((((((...)))))).))..)))).)))))..----- ( -48.30, z-score = -3.36, R) >droSec1.super_1 12368341 110 + 14215200 CCAGCGGCGGUUGCCAGCGAG-GAAGCGGAAGUCAUCGAGAUC---GACGACGGCGAUGCGGCCGCAAAUGCAGAGGUGGCGGCUAUCAACGAUGGCAGCAACACCAGCAGGAG----- ((.(((((.((((((.((...-...))....(((.(((....)---)).)))))))..)).)))))...(((...((((((.((((((...)))))).))..)))).)))))..----- ( -46.00, z-score = -2.79, R) >droYak2.chr2R 6807052 110 - 21139217 CCAGCGGCGGUUGCCAGCGAG-GAGGCAGAGGUCAUCGAGAUU---GACGAUGGCGAUGCGGCUGCAAAUGCAGAGGUGGCGGCCAUCAAUGAUGGCAACAAUGCCAGCAGGAG----- ((.(((((.(((((((.((..-((.(((...(((((((.....---..)))))))..)))((((((....((....)).)))))).))..)).)))))))...))).)).))..----- ( -45.60, z-score = -2.68, R) >droEre2.scaffold_4845 9053935 110 + 22589142 CCAGCGGCGGUUGCCAGCGAG-GAAGCAGAGGUCAUUGAGAUU---GACGAUGGCGAUGCGGCCGCAAAUGCGGAAGUGGCGGCCAUCAAUGAUGGCAGCAAUACCAGCAGGAG----- ((.((((..(((((((.((..-((.(((...(((((((.....---..)))))))..)))((((((....((....)).)))))).))..)).)))))))....)).)).))..----- ( -46.00, z-score = -3.24, R) >droAna3.scaffold_13266 2410363 110 - 19884421 -CCGGCGGAGCAGGUGGAGGCAGAGCCCGAGGUUAUCGAAAUC---GAUGAUGGCGAUGCGGCCGCCAAUGCCGAGGUGGCAGCCAUCAAUGAUGGUAGCAGUUCCAGUCGGAG----- -((((((((((.(((((..(((..(((....(((((((....)---)))))))))..)))..)))))..((((.....))))((((((...))))))....))))).)))))..----- ( -53.20, z-score = -4.54, R) >dp4.chr3 5936869 98 + 19779522 -------------CCGGCCAGCGAGUCGGAAGUGAUCGAGAUC---GAUGACGGCGAUGCCGCUGCCAAUGCCGAGGUGGCCGCGAUCAACGAUGGCAGCAACACAAACAGGCG----- -------------..(((.((((.((((...((.((((....)---))).))..))))..)))))))...(((...(((((.((.(((...))).)).))..))).....))).----- ( -37.50, z-score = -1.16, R) >droPer1.super_2 6138024 98 + 9036312 -------------CCGGCCAGCGAGUCGGAAGUGAUCGAGAUC---GAUGACGGCGAUGCCGCUGCCAAUGCCGAGGUGGCCGCGAUCAACGAUGGCAGCAACACAAACAGGCG----- -------------..(((.((((.((((...((.((((....)---))).))..))))..)))))))...(((...(((((.((.(((...))).)).))..))).....))).----- ( -37.50, z-score = -1.16, R) >droMoj3.scaffold_6496 20186305 98 - 26866924 -------------------CCGGCCAACGAGAUCAUCGAGAUCGGCGAUGAUGGCGAUGCGGAGGCCAAUGCCGAGGUGGCCGCCAUCAAUGAUGGCACCGGG--CAGACGGGACGACG -------------------((((((..((..(((((((.......)))))))..))...(((.((((...((....))))))((((((...)))))).)))))--)...)))....... ( -39.20, z-score = -1.23, R) >droVir3.scaffold_12875 991924 95 - 20611582 -------------------CCCGCCAACGAGGUAAUCGAAAUU---GAUGACGGUGAUGCUGCUGCCAAUGCCGAAGUGGCUGCCAUCAAUGAUGGCAGCAGC--CAGACUGGUCGCCG -------------------...(((.....))).((((....)---)))..(((((((.(((((((....(((.....)))(((((((...))))))))))).--)))....))))))) ( -33.60, z-score = -1.42, R) >droGri2.scaffold_15245 17936464 95 + 18325388 -------------------UCCGCCAACGAGGUGAUCGAGAUC---GAUGACGGCGAUGCUGCCGCCAAUGCUGAAGUGGCCGCCAUCAAUGAUAGCAGCAAU--CAGACGGGACGCCG -------------------..((((.....))))((((....)---)))..(((((.((((((.((((.(.....).))))....(((...))).)))))).(--(......))))))) ( -27.60, z-score = 0.72, R) >consensus _C_G__G__G__GCCAGCGAG_GAAGCCGAGGUCAUCGAGAUC___GAUGACGGCGAUGCGGCCGCCAAUGCCGAGGUGGCGGCCAUCAAUGAUGGCAGCAACACCAGCAGGAG_____ ............................(..((.((((...........(....)(((((.(((((..........))))).)).)))..)))).))..)................... (-13.18 = -12.78 + -0.39)

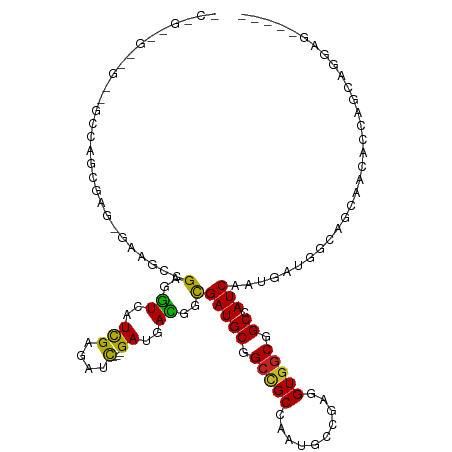
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:25 2011