
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,855,102 – 14,855,254 |
| Length | 152 |
| Max. P | 0.591536 |

| Location | 14,855,102 – 14,855,254 |
|---|---|
| Length | 152 |
| Sequences | 6 |
| Columns | 170 |
| Reading direction | reverse |
| Mean pairwise identity | 65.63 |
| Shannon entropy | 0.60871 |
| G+C content | 0.36772 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -16.73 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14855102 152 - 21146708 ------UAUAUAACUGAAGGCCUAUGUAUG--UUU-AAACUCAUAUGAGUGUUUC-UCAAUUUUUUUUUUAUUAUUGGUGACAC-UUUAUGUCUUUUAUUGCCACCGCAAUCAUGUGGAUACGUGAUGGUA------UGGCAUUA-GGCAAAGGCCACUAUCAUGGUCAA ------............(((((..(((((--...-.....)))))(((((((.(-.((((..(......)..))))).)))))-))..(((((.....(((((..((.(((((((....))))))).)).------)))))..)-)))).))))).............. ( -39.20, z-score = -1.77, R) >droSim1.chr2R 13553784 151 - 19596830 ------UAGAUAACUGAAGGCCUGUGUAUA--UUU-AAACUCAUAUGAGUGUUUC-UCAA-UUUUUUUUUAUUAUUGGUGACAC-UUUAUGUCUUUUAUUGCCACCGCAAUCAUGUGGAUACGUGAUGGUA------UGGCAUUA-GGCAAAGGCCACUAUCAUGGUCAA ------..(((...(((.(((((.(((...--...-.....((((.(((((((.(-.(((-(..(.....)..))))).)))))-))))))....(((.(((((..((.(((((((....))))))).)).------))))).))-)))).)))))....)))..))).. ( -40.20, z-score = -1.85, R) >droSec1.super_1 12361101 151 - 14215200 ------UAGAUAACUGAAGACAUACGUAUA--UUU-AAACUCAUCUGAGUGUUUC-UAAA-UAUUUUUUUAUUAUUGGUGACAC-UUUAUGUCUUUUAUUGCCACCGCAAUCAUGUGGAUACGUGAUGGUA------UGGCAUUA-GGCAAAGGCCACUAUCAUGGUCAA ------..((((...(((((((((...(((--(((-(.((((....)))).....-))))-)))...........((....)).-..)))))))))((.(((((..((.(((((((....))))))).)).------))))).))-(((....)))..))))........ ( -37.30, z-score = -1.71, R) >droYak2.chr2R 6799579 156 + 21139217 ------UAGAAAUAUAAUGGCCUAUAAAUA--UUUCGAAUUCAUAUGAGUGUUUC-UCAA---UUUUGUCAUUACUGGUGACAC-UUUACGUCUUUUAUUGCCACCACAAUCAUGUGGAUACGUGGUGGUACGAGUGUGGCAUUA-GGUAAAGGCCACUAUCAUGGUCAA ------..((.(((((.((((((.((..((--.............((((.....)-))).---...((((((.(((.(((((..-.....))).......((((((((.(((.....)))..)))))))))).))))))))).))-..)).)))))).))).))..)).. ( -41.50, z-score = -1.54, R) >droEre2.scaffold_4845 9046669 133 - 22589142 ------UAGAAAACUGGUGGCCUAUCCAUA--UCUUGAAUUCAUAUAAGUGCCAC-UCAA---UUUUUUCAUUAUUGGUGACAC-UUUAUGUCUUUUAUUGCCACCACAAUCAUGUGGAUACGUGAUGGUA------UGGCGUCAUGGUCAA------------------ ------...........(((((...(((((--((..(((..((((.(((((.(((-.(((---(.........))))))).)))-))))))..))).((..(..(((((....)))))....)..))))))------)))......))))).------------------ ( -32.80, z-score = -0.92, R) >droGri2.scaffold_15245 17928425 152 - 18325388 UAUGCAUAUGUAUGUGGGUGUAUGUGUAUAAGAUAUAUGCUUAAAUUAAUGUUUUAUUAAACGCGCUGAGACU-CUGGCGAUUCAUCUUCAUUCCUUGCCAUUGCCAUUGGCUAUUGGCUGCUCAGACCAACAGAGACAGAGACAGAGCAGAG----------------- (((((((((((((....))))))))))))).......(((((.......((((((..........(((((...-.((((((..............))))))..((((........))))..))))).......))))))......)))))...----------------- ( -33.89, z-score = 1.34, R) >consensus ______UAGAUAACUGAAGGCCUAUGUAUA__UUU_AAACUCAUAUGAGUGUUUC_UCAA_U_UUUUUUUAUUAUUGGUGACAC_UUUAUGUCUUUUAUUGCCACCACAAUCAUGUGGAUACGUGAUGGUA______UGGCAUUA_GGCAAAGGCCACUAUCAUGGUCAA ...............((((((.................((((....)))).........................((....)).......))))))..(((((.(((((....))))).....(((((.((......)).))))).))))).(((((......))))).. (-16.73 = -17.03 + 0.30)
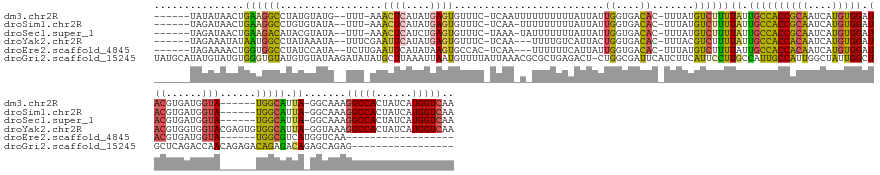
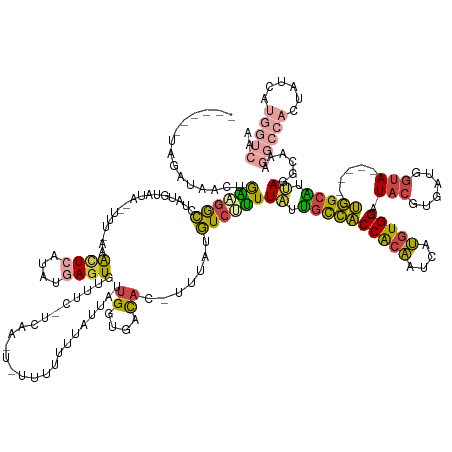
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:23 2011