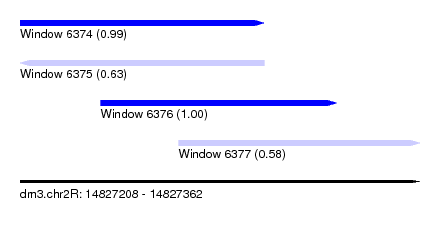
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,827,208 – 14,827,362 |
| Length | 154 |
| Max. P | 0.998502 |

| Location | 14,827,208 – 14,827,302 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.43820 |
| G+C content | 0.47401 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -14.88 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
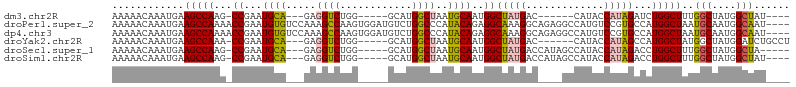
>dm3.chr2R 14827208 94 + 21146708 AAAAACAAAUGAAGCCAAG-CCGAAUGCA---GAGGUCUGG-----GCAUGGCUAAUGCAAUGGCUAUGAC------CAUACCAUAGAUCUGGCUUUGGCUAUGGCUAU---- ............(((((((-(((((.(((---(((((.(((-----.((((((((......)))))))).)------)).))).....))).))))))))).)))))..---- ( -37.40, z-score = -3.93, R) >droPer1.super_2 6104313 109 + 9036312 AAAAACAAAUGAAGCCAAAACCGAAUGUGUCCAAAGCCAAGUGGAUGUCUGGCCCAUACAGAGGCAAAGGCAGAGGCCAUGUCCGUGCCAUGGCUAAUGCAAUGGCAAU---- .............((((...((...(((((.....((((..(....)..))))..)))))..)).....(((..(((((((.......)))))))..)))..))))...---- ( -31.00, z-score = -0.68, R) >dp4.chr3 5903294 109 + 19779522 AAAAACAAAUGAAGCCAAAACCGAAUGUGUCCAAAGCCAAGUGGAUGUCUGGCCCAUACAGAGGCAAAGGCAGAGGCCAUGUCCGUGCCAUGGCUAAUGCAAUGGCAAU---- .............((((...((...(((((.....((((..(....)..))))..)))))..)).....(((..(((((((.......)))))))..)))..))))...---- ( -31.00, z-score = -0.68, R) >droYak2.chr2R 6771338 98 - 21139217 AAAAACAAAUGAAGCCAAA-CCGAAUGCA---GAGGUCUGG-----GCAUGGCUAAUGCAAUGGCUAUGAC------CAUACCAUAGCCAUGGCUAUGGCUAUGGAUCUGCCU ...................-......(((---(((((((((-----.(((.((....)).))).))).)))------)...((((((((((....)))))))))).))))).. ( -38.00, z-score = -3.72, R) >droSec1.super_1 12333503 99 + 14215200 AAAAACAAAUGAAGCCAAG-CCGAAUGCA---GAGGUCUGG-----GCAUGGCUAAUGCAAUGGCUAUGACCAUAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUA----- ............(((((((-(((((.((.---.((((((((-----((((.....)))).((((((((....)))))))).))..)))))).))))))))).))))).----- ( -44.90, z-score = -5.24, R) >droSim1.chr2R 13526370 100 + 19596830 AAAAACAAAUGAAGCCAAG-CCGAAUGCA---GAGGUCUGG-----GCAUGGCUAAUGCAAUGGCUAUGACCAUAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUAU---- ............(((((((-(((((.((.---.((((((((-----((((.....)))).((((((((....)))))))).))..)))))).))))))))).)))))..---- ( -44.90, z-score = -5.24, R) >consensus AAAAACAAAUGAAGCCAAA_CCGAAUGCA___GAGGUCUGG_____GCAUGGCUAAUGCAAUGGCUAUGAC____GCCAUACCAUAGACAUGGCUAUGGCUAUGGCUAU____ .............((((.................(((.(((......((((((((......)))))))).......))).))).......((((....))))))))....... (-14.88 = -16.60 + 1.73)
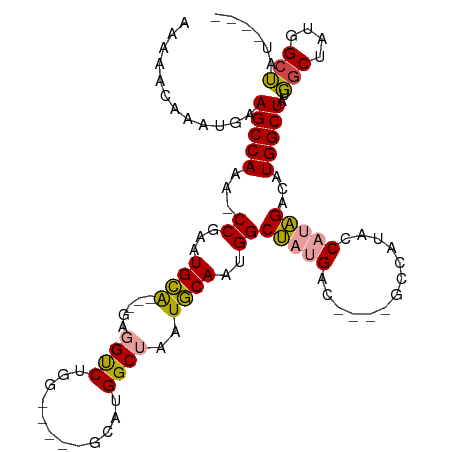
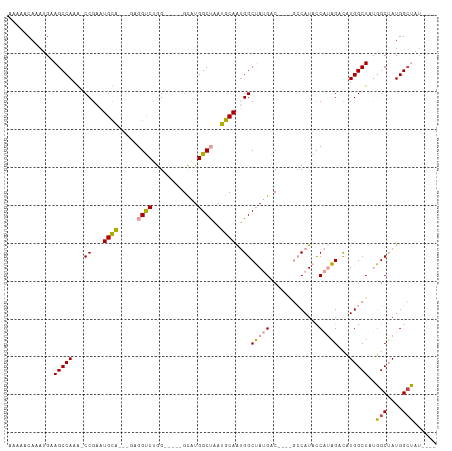
| Location | 14,827,208 – 14,827,302 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.43820 |
| G+C content | 0.47401 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -12.04 |
| Energy contribution | -11.72 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14827208 94 - 21146708 ----AUAGCCAUAGCCAAAGCCAGAUCUAUGGUAUG------GUCAUAGCCAUUGCAUUAGCCAUGC-----CCAGACCUC---UGCAUUCGG-CUUGGCUUCAUUUGUUUUU ----..(((((.((((.((..(((((((..((((((------((....((....))....)))))))-----).)))..))---))..)).))-)))))))............ ( -30.50, z-score = -2.45, R) >droPer1.super_2 6104313 109 - 9036312 ----AUUGCCAUUGCAUUAGCCAUGGCACGGACAUGGCCUCUGCCUUUGCCUCUGUAUGGGCCAGACAUCCACUUGGCUUUGGACACAUUCGGUUUUGGCUUCAUUUGUUUUU ----...(((((.((....)).)))))(((((...((((...(((..((..(((....(((((((........))))))).)))..))...)))...))))...))))).... ( -31.30, z-score = -0.52, R) >dp4.chr3 5903294 109 - 19779522 ----AUUGCCAUUGCAUUAGCCAUGGCACGGACAUGGCCUCUGCCUUUGCCUCUGUAUGGGCCAGACAUCCACUUGGCUUUGGACACAUUCGGUUUUGGCUUCAUUUGUUUUU ----...(((((.((....)).)))))(((((...((((...(((..((..(((....(((((((........))))))).)))..))...)))...))))...))))).... ( -31.30, z-score = -0.52, R) >droYak2.chr2R 6771338 98 + 21139217 AGGCAGAUCCAUAGCCAUAGCCAUGGCUAUGGUAUG------GUCAUAGCCAUUGCAUUAGCCAUGC-----CCAGACCUC---UGCAUUCGG-UUUGGCUUCAUUUGUUUUU .(((.((.(((((.(((((((....)))))))))))------)))...)))...(((..(((((.((-----((((....)---)).....))-).))))).....))).... ( -35.90, z-score = -2.91, R) >droSec1.super_1 12333503 99 - 14215200 -----UAGCCAUAGCCAAAGCCAGGUCUAUGGUAUGGCUAUGGUCAUAGCCAUUGCAUUAGCCAUGC-----CCAGACCUC---UGCAUUCGG-CUUGGCUUCAUUUGUUUUU -----.(((((.((((((.((.((((((..((((((((((((((....))))......)))))))))-----).)))))).---.)).)).))-)))))))............ ( -41.40, z-score = -4.27, R) >droSim1.chr2R 13526370 100 - 19596830 ----AUAGCCAUAGCCAAAGCCAGGUCUAUGGUAUGGCUAUGGUCAUAGCCAUUGCAUUAGCCAUGC-----CCAGACCUC---UGCAUUCGG-CUUGGCUUCAUUUGUUUUU ----..(((((.((((((.((.((((((..((((((((((((((....))))......)))))))))-----).)))))).---.)).)).))-)))))))............ ( -41.40, z-score = -4.23, R) >consensus ____AUAGCCAUAGCCAAAGCCAGGGCUAUGGUAUGGC____GUCAUAGCCAUUGCAUUAGCCAUGC_____CCAGACCUC___UGCAUUCGG_CUUGGCUUCAUUUGUUUUU .......(((((.((((.((......)).)))))))))....((((...((..((((..((...((.......))...))....))))...))...))))............. (-12.04 = -11.72 + -0.33)
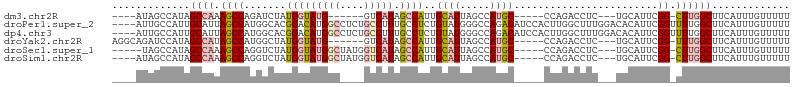
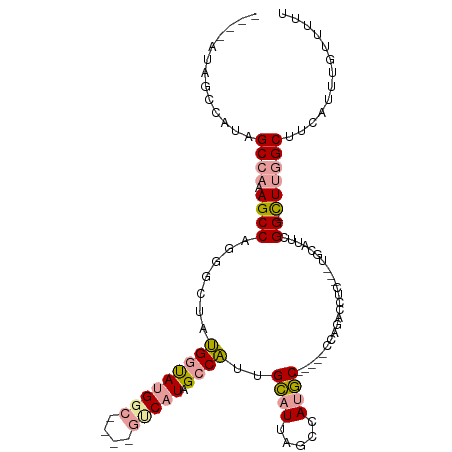
| Location | 14,827,239 – 14,827,330 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Shannon entropy | 0.30663 |
| G+C content | 0.52012 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -27.71 |
| Energy contribution | -30.53 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14827239 91 + 21146708 GUCUGGGCAUGGCUAAUGCAAUGGCUAUGA------CCAUACCAUAGAUCUGGCUUUGGCUAUGGCUAUGGCUAUGGAUCU----GGCUCUGACUCUGGCU------------ (((.(((((((((((......)))))))((------(((((((((((.((((((....)))).)))))))).))))).)).----.)))).))).......------------ ( -34.90, z-score = -2.84, R) >droYak2.chr2R 6771369 107 - 21139217 GUCUGGGCAUGGCUAAUGCAAUGGCUAUGA------CCAUACCAUAGCCAUGGCUAUGGCUAUGGAUCUGCCUCUCGGAGUCAGAGGCUCUGGCUCUGGCUUUUGCUUUGGCU (((..((((.(((.........(((...((------((((((((((((....))))))).))))).)).)))...(((((((((.....))))))))))))..))))..))). ( -50.30, z-score = -3.95, R) >droSec1.super_1 12333534 91 + 14215200 GUCUGGGCAUGGCUAAUGCAAUGGCUAUGACCAUAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUAUGG------AUCU----GGCUCUGACUCUGGCU------------ (((.(((((((((((......)))))))((((((((((((((((.((......)).))).)))))))))))------.)).----.)))).))).......------------ ( -39.00, z-score = -3.60, R) >droSim1.chr2R 13526401 97 + 19596830 GUCUGGGCAUGGCUAAUGCAAUGGCUAUGACCAUAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUAUGGCUGUGGAUCU----GGCUCUGACUCUGGCU------------ (((.(((((((((((......)))))))(((((((((((((((((((.((.......)))))))).))))))))))).)).----.)))).))).......------------ ( -46.00, z-score = -4.66, R) >consensus GUCUGGGCAUGGCUAAUGCAAUGGCUAUGA______CCAUACCAUAGACCUGGCUUUGGCUAUGGCUAUGGCU_UGGAUCU____GGCUCUGACUCUGGCU____________ (((.(((((((((((......)))))))..((((((((((((((.((......)).))).)))))))))))...............)))).)))................... (-27.71 = -30.53 + 2.81)

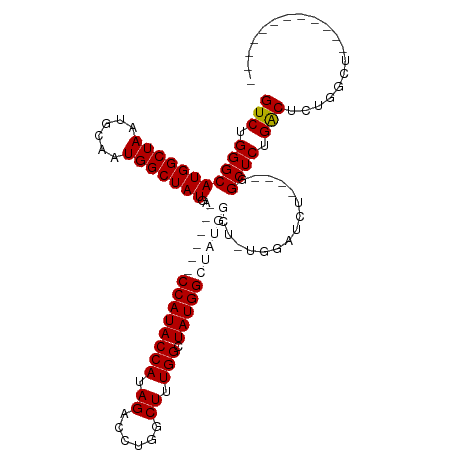
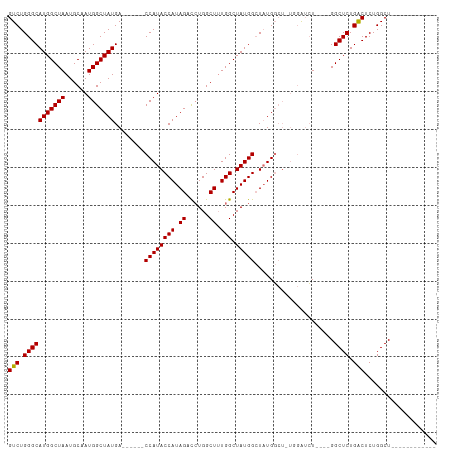
| Location | 14,827,269 – 14,827,362 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Shannon entropy | 0.28368 |
| G+C content | 0.53540 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.45 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14827269 93 + 21146708 ---CCAUACCAUAGAUCUGGCUUUGGCUAUGGCUAUG----------------GCUAUGGAUCUGGCUCUGACUCUGGCUAACUAAGAUCCGUAACCCGCUCGCUUCGAUGC ---(((((((((((.((.......)))))))).))))----------------).(((((((((((((........)))).....)))))))))....(((((...))).)) ( -26.70, z-score = -1.24, R) >droYak2.chr2R 6771399 109 - 21139217 ---CCAUACCAUAGCCAUGGCUAUGGCUAUGGAUCUGCCUCUCGGAGUCAGAGGCUCUGGCUCUGGCUUUUGCUUUGGCUAACUAAGAUCCGUAACCCGCUCGCUCCGAUGC ---.....(((((((....))))))).(((((((((((((((.......))))))..(((((..(((....)))..)))))....)))))))))....(((((...))).)) ( -42.30, z-score = -2.81, R) >droSec1.super_1 12333567 90 + 14215200 UAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUAU----------------------GGAUCUGGCUCUGACUCUGGCUAACUAAGAUCCGUAACCCGCUCGCUCCGAUGC ..(((((((((.((......)).))).))))))(((----------------------((((((((((........)))).....)))))))))....(((((...))).)) ( -26.80, z-score = -1.45, R) >droSim1.chr2R 13526434 96 + 19596830 UAGCCAUACCAUAGACCUGGCUUUGGCUAUGGCUAUG----------------GCUGUGGAUCUGGCUCUGACUCUGGCUAACUAAGAUCCGUAACCCGCUCGCUCCGAUGC ((((((((((((((.((.......)))))))).))))----------------)))).((((((((((........)))).....)))))).......(((((...))).)) ( -32.80, z-score = -2.17, R) >consensus ___CCAUACCAUAGACCUGGCUUUGGCUAUGGCUAUG________________GCU_UGGAUCUGGCUCUGACUCUGGCUAACUAAGAUCCGUAACCCGCUCGCUCCGAUGC .((((((((((.((......)).))).)))))))........................((((((((((........)))).....)))))).......(((((...))).)) (-20.20 = -21.45 + 1.25)

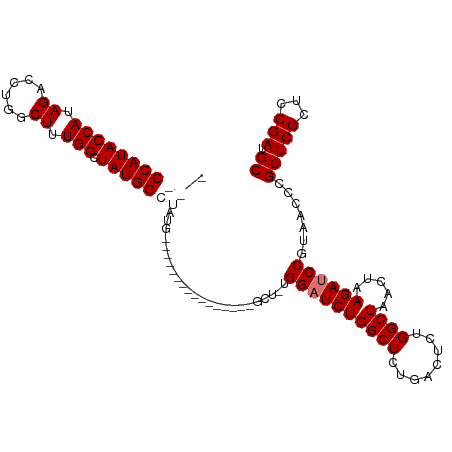
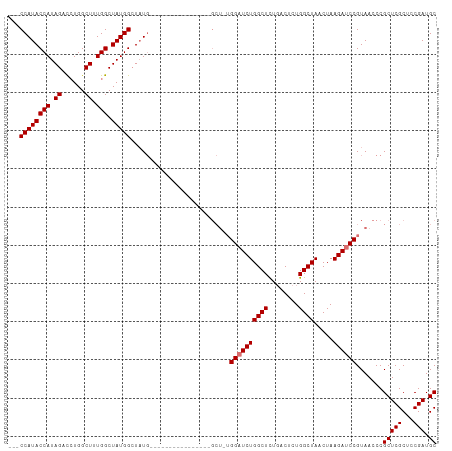
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:16 2011