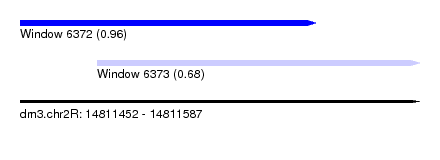
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,811,452 – 14,811,587 |
| Length | 135 |
| Max. P | 0.964966 |

| Location | 14,811,452 – 14,811,552 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Shannon entropy | 0.30025 |
| G+C content | 0.57024 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
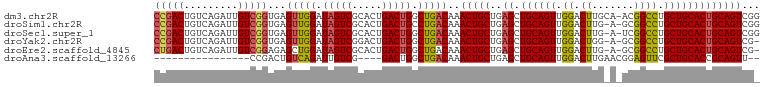
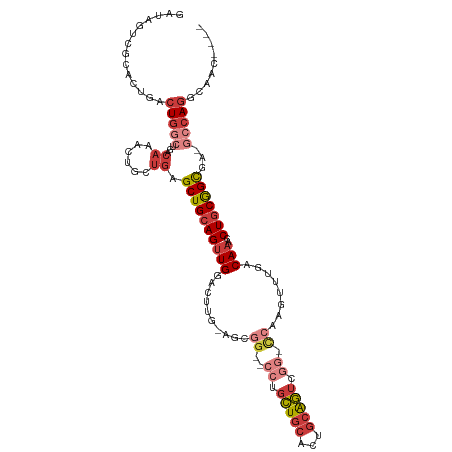
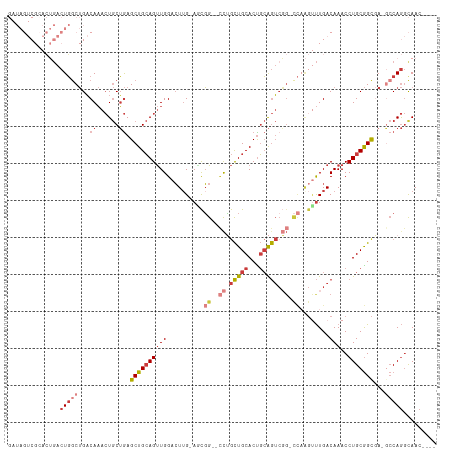
>dm3.chr2R 14811452 100 + 21146708 CCGACUGUCAGAUUGUCGGUGAGUUGGAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUGCA-ACGGCCUGCUGCACUGCAGUCGG (((((((((((.((((((((.(((..(.........)..))).)))))))).)))...((.((((((.((.((....-..)))).)))))))))))))))) ( -44.20, z-score = -3.08, R) >droSim1.chr2R 13510510 99 + 19596830 CCGACUGUCAGAUUGUCGGUGAGUUGGAUAGUCGCACUGACUGCCUGACAAACUGCUGAGCUGCAGUUGGACUUG-A-GCGGCCUGCUGCACUGCAGUCGG (((((((.(((...(((.....((..(.(((((.....))))).)..)).((((((......)))))).)))...-.-((((....)))).)))))))))) ( -41.30, z-score = -2.28, R) >droSec1.super_1 12317587 99 + 14215200 CCGACUGUCAGAUUGUCGGUGAGUUGGAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG-A-UCGGCCUGCUGCACUGCAGUCGG (((((((((((.((((((((.(((..(.........)..))).)))))))).)))...((.((((((.((.((..-.-..)))).)))))))))))))))) ( -44.00, z-score = -3.37, R) >droYak2.chr2R 6755042 98 - 21139217 CCGACUGUCAGAUUGUCGGUGAGUUGGAUAGUCGGACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUGG-A-GCGGCCUGCUGCACUGCAGUCG- .((((((((((.((((((((.(((..(.........)..))).)))))))).)))...((.((((((.((.(((.-.-.))))).)))))))))))))))- ( -42.20, z-score = -2.73, R) >droEre2.scaffold_4845 9003057 98 + 22589142 CUGACUGUCAGAUUGUCGGAGAGCUGGAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG-A-GCGGCCUGCUGCACUGCAGUCG- ..(((((.(((...(((....(((..(.(((.....))).)..)))....((((((......)))))).)))...-.-((((....)))).)))))))).- ( -36.20, z-score = -0.74, R) >droAna3.scaffold_13266 2357264 79 - 19884421 ----------------CCGACUGUCAGAUUGUCG----GACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUGAACGGAGUUCGCUGCACCGCAGUU-- ----------------..(((((((((.((((((----(.....))))))).)))....(.((((((.((((((.....)))))))))))).)))))))-- ( -29.40, z-score = -1.83, R) >consensus CCGACUGUCAGAUUGUCGGUGAGUUGGAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG_A_GCGGCCUGCUGCACUGCAGUCG_ (((((.........)))))...(((((.(((((.....))))).)))))..(((((..((.((((((.((.((.......)))).)))))))))))))... (-28.80 = -29.08 + 0.28)
| Location | 14,811,478 – 14,811,587 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Shannon entropy | 0.45925 |
| G+C content | 0.56967 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -18.13 |
| Energy contribution | -20.11 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14811478 109 + 21146708 GAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUGCAACGG--CCUGCUGCACUGCAGUCGGGCCAAGUUUGACAAACCUGCGGCUA-GCCAGGCAAC---- .......((.(((.(((((((.((...(((......)))((..(((((.....((--((((((((...)))).)))))))))))..)).....))))))))-).)))))...---- ( -44.80, z-score = -2.01, R) >droSim1.chr2R 13510536 108 + 19596830 GAUAGUCGCACUGACUGCCUGACAAACUGCUGAGCUGCAGUUGGACUUG-AGCGG--CCUGCUGCACUGCAGUCGGGCCAAGUUUGACAAACCUGCGGCGA-GCCAGGCAAC---- ...((((.....))))(((((.......(((..((((((((..(((((.-...((--((((((((...)))).)))))))))))..)).....)))))).)-)))))))...---- ( -44.91, z-score = -1.97, R) >droSec1.super_1 12317613 108 + 14215200 GAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG-AUCGG--CCUGCUGCACUGCAGUCGGGCCAAGUUUGACAAACCUGCGGCGA-GCCAGGCAAC---- .......((.(((.((.((((.((...(((......)))((..(((((.-...((--((((((((...)))).)))))))))))..)).....)))))).)-).)))))...---- ( -43.10, z-score = -1.78, R) >droYak2.chr2R 6755068 108 - 21139217 GAUAGUCGGACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUGG-AGCGG--CCUGCUGCACUGCAGUCGG-CCAAGUUUGACAAACCUGCGGCGAUACCAGGCAAC---- ....((((((((...((((((((.((((((......)))))).....((-.((((--....)))).))...)))))-)))))))))))...((((.(......)))))....---- ( -42.50, z-score = -2.11, R) >droEre2.scaffold_4845 9003083 107 + 22589142 GAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG-AGCGG--CCUGCUGCACUGCAGUCGG-CCAAGUUUGACAAACCUGCGGCGA-GCCAGGCAAC---- .......((.(((.((.((((.((...(((......)))((..((((((-.((.(--.((((......)))).).)-)))))))..)).....)))))).)-).)))))...---- ( -42.40, z-score = -1.27, R) >droAna3.scaffold_13266 2357282 88 - 19884421 ------------GACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG-AACGGAGUUCGCUGCACCGCAGU----------UUGACAAACCUGCGGUGA-GCCAGGUAAC---- ------------..((((((....((((((......))))))((((((.-....))))))....((((((((.----------((....)).)))))))))-))))).....---- ( -37.50, z-score = -3.61, R) >droMoj3.scaffold_6496 20113094 103 - 26866924 ----------GGCAAUCGCAGUCACAGU-CGAAGCUGGAGCUGGAGUUGGAGUUGUUGCAAUUGCACUUGGAGCUG-UUUGGCCUGACAAACCUGCAGCAACAGCAA-CAACAAAA ----------..((((..((((..((((-....))))..))))..))))..((((((((..((((...((.((..(-((((......))))))).))))))..))))-)))).... ( -35.00, z-score = -0.75, R) >consensus GAUAGUCGCACUGACUGGCUGACAAACUGCUGAGCUGCAGUUGGACUUG_AGCGG__CCUGCUGCACUGCAGUCGG_CCAAGUUUGACAAACCUGCGGCGA_GCCAGGCAAC____ ..............(((((...(.((((((......)))))).)................((((((.....(((((.......))))).....))))))...)))))......... (-18.13 = -20.11 + 1.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:13 2011