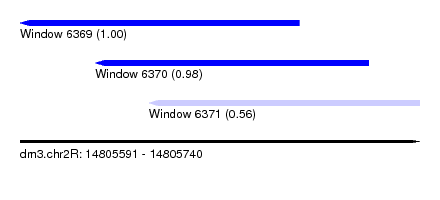
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,805,591 – 14,805,740 |
| Length | 149 |
| Max. P | 0.998572 |

| Location | 14,805,591 – 14,805,695 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.62965 |
| G+C content | 0.34626 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -10.53 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998572 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 14805591 104 - 21146708 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCAAACCAAAAAAUCAAC-AAAACCACGAAACAAA-- ....((((((((.((-((.((.....)).)))).)))-)))))((..(((-(((....))))))..)).......................-................-- ( -21.50, z-score = -3.45, R) >droSim1.chr2R 13504228 105 - 19596830 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCAAACCAAAAAAUCAACAAAAACCACGAAAUAAA-- ....((((((((.((-((.((.....)).)))).)))-)))))((..(((-(((....))))))..))........................................-- ( -21.50, z-score = -3.41, R) >droSec1.super_1 12311707 104 - 14215200 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCAAACCAAAAAAUCAAC-AAAACCACGAAAUAAA-- ....((((((((.((-((.((.....)).)))).)))-)))))((..(((-(((....))))))..)).......................-................-- ( -21.50, z-score = -3.49, R) >droYak2.chr2R 6749010 103 + 21139217 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCAAGCCAAAAAAUCAAC-AAAACCAC-GAAUAAA-- ....((((((((.((-((.((.....)).)))).)))-)))))((..(((-(((....))))))..)).......................-........-.......-- ( -21.50, z-score = -2.91, R) >droEre2.scaffold_4845 8997206 103 - 22589142 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCGAACCAAAAAAUCAAC-AAAACCAC-AAGUAAA-- ....((((((((.((-((.((.....)).)))).)))-)))))((..(((-(((....))))))..)).......................-........-.......-- ( -21.50, z-score = -2.99, R) >droAna3.scaffold_13266 2351539 80 + 19884421 AAAUUGCGCCCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAC-AUUCAUGAAUUUACACGCCAAA------CAAAAAGCCA--------------------- .....((((.((.((-((.((.....)).)))).)).-))))(((((...-.)))))................------..........--------------------- ( -16.50, z-score = -0.88, R) >droWil1.scaffold_180697 2383016 106 - 4168966 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGAGGUGCUUGAAUAA-AUGCAUGAGUUUACA--AAAAUAAAAAAUAAAAACAAAUUAACAUUAUAUGGAAAAAAA ...(((.(((((...-..(((((.(((.(((((......))))).))).)-)))).)))))...))--)......................................... ( -15.90, z-score = -0.33, R) >droMoj3.scaffold_6496 20104274 87 + 26866924 AAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCACAUAUAAGAUGCAUGAAUUUAUACAACAACAACGGGCAACAACAAG--------------------- ....((((((((.((-((.((.....)).)))).)))-)))))..................................(....)......--------------------- ( -21.20, z-score = -2.69, R) >droGri2.scaffold_15245 17876968 103 - 18325388 AAAUUGCGCUCAAUG-CCGAAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUGCAUUAAUUUACAC-ACAAAAAUCAACAAUGACAAGCCAAAUGGUUUCUAUUAA--- ....((((((((.((-((...........)))).)))-)))))((..(((-((......)))))..)-).................(((((....))))).......--- ( -25.00, z-score = -2.74, R) >anoGam1.chr3R 22651886 105 - 53272125 UAGUAGCUGUCGGUGUUUUUUUUUGUUUAGAAAACAA-AAACAGGAAUAA-GUUCAAGGGGUAACAGAACAAACCAAACCAAAAAGUAAAGGUAAAUAGAAGAACAA--- ......((((...((((((((.......)))))))).-..))))......-((((..((.((......))...))..(((..........)))........))))..--- ( -17.10, z-score = -0.98, R) >consensus AAAUUGCGCUCAAUG_UCGCAUUAAUUGAGGCAAUGA_GCGCAUGAAUAA_AUUCAUGAAUUUACACACCAAACCAAACCAAAAAAUCAAC_AAAACCAC_AAAUAAA__ ..(((((.((((((..........)))))))))))......(((((((...))))))).................................................... (-10.53 = -11.25 + 0.72)

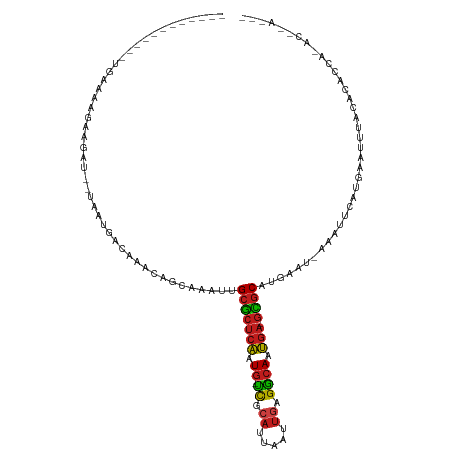
| Location | 14,805,619 – 14,805,721 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.54408 |
| G+C content | 0.36341 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.44 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14805619 102 - 21146708 ------------UGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUUCAUGAAUUUACACACCAGACCAAAC- ------------......(.((((--(.((((.......((...((((((((.((-((.((.....)).)))).)))-)))))))...-....)))).))))).)..............- ( -24.04, z-score = -3.29, R) >droSim1.chr2R 13504257 102 - 19596830 ------------UGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUUCAUGAAUUUACACACCAGACCAAAC- ------------......(.((((--(.((((.......((...((((((((.((-((.((.....)).)))).)))-)))))))...-....)))).))))).)..............- ( -24.04, z-score = -3.29, R) >droSec1.super_1 12311735 102 - 14215200 ------------UGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUUCAUGAAUUUACACACCAGACCAAAC- ------------......(.((((--(.((((.......((...((((((((.((-((.((.....)).)))).)))-)))))))...-....)))).))))).)..............- ( -24.04, z-score = -3.29, R) >droYak2.chr2R 6749037 102 + 21139217 ------------UGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUUCAUGAAUUUACACACCAGACCAAGC- ------------......(.((((--(.((((.......((...((((((((.((-((.((.....)).)))).)))-)))))))...-....)))).))))).)..............- ( -24.04, z-score = -2.75, R) >droEre2.scaffold_4845 8997233 102 - 22589142 ------------UGAAAAUAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUUCAUGAAUUUACACACCAGACCGAAC- ------------........((((--(.((((.......((...((((((((.((-((.((.....)).)))).)))-)))))))...-....)))).)))))................- ( -23.74, z-score = -3.07, R) >droAna3.scaffold_13266 2351548 108 + 19884421 AGUCUUGCGGCUUGAAAGGAAGAU--UAAUGACAAC-AGCAAAUUGCGCCCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-ACAUUCAUGAAUUUACACGCCAAAC------ ........(((.((..........--..........-........((((.((.((-((.((.....)).)))).)).-))))(((((.-...))))).......)).)))....------ ( -23.20, z-score = -0.09, R) >dp4.chr3 5881307 79 - 19779522 ------------GGGGUUGAAGAU--UAAUGACCAACAGAAAUUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCUUGAAU-AAAUACA------------------------ ------------..((((......--....))))...........(((((((.((-((.((.....)).)))).)))-))))......-.......------------------------ ( -21.80, z-score = -2.61, R) >droPer1.super_2 6082556 79 - 9036312 ------------GGGGUUGAAGAU--UAAUGACAAACAGAAAUUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCUUAAAU-AAAUACA------------------------ ------------(..(((((...)--))))..)............(((((((.((-((.((.....)).)))).)))-))))......-.......------------------------ ( -19.70, z-score = -2.54, R) >droWil1.scaffold_180697 2383044 104 - 4168966 ------------GCUUAAGAAGAU--UAAUGACAAACAACAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGAGGUGCUUGAAU-AAAUGCAUGAGUUUACAAAAAUAAAAAAUAA ------------(((((.......--....((((...................))-))(((((.(((.(((((......))))).)))-.))))).)))))................... ( -17.31, z-score = -0.30, R) >droVir3.scaffold_12875 919027 79 + 20611582 ------------UGAAAAGAAGAU--UAAUGACAAACGACAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAU-UUACGCA------------------------ ------------............--..................((((((((.((-((.((.....)).)))).)))-))))).....-.......------------------------ ( -20.30, z-score = -2.21, R) >droMoj3.scaffold_6496 20104295 92 + 26866924 ------------UGCAAAGAAGAU--UAAUGACAACUGACAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCACAUAUAAGAUGCAUGAAUUUAUACA------------ ------------........((((--(.(((.((.((.......((((((((.((-((.((.....)).)))).)))-)))))......)).))))).))))).....------------ ( -25.02, z-score = -2.85, R) >droGri2.scaffold_15245 17877005 92 - 18325388 ------------AAAAAAGAAGAU--UAAUGACAAAUGACAAAUUGCGCUCAAUG-CCGAAUUAAUUGAGGCAAUGA-GCGCAUGAAU-AAAUGCAUUAAUUUACACAC----------- ------------......(.((((--(((((.((..........((((((((.((-((...........)))).)))-))))).....-...))))))))))).)....----------- ( -27.57, z-score = -5.93, R) >anoGam1.chr3R 22651914 105 - 53272125 ------------GGAAGAGAAGAUAAAAACAACGUUCCGUUAGUAGCUGUCGGUGUUUUUUUUUGUUUAGAAAACAA-AAACAGGAAU-AAGUUCAAGGGGUAACAGAACAAACCAAAC- ------------.....................((((.((((.(..((((...((((((((.......)))))))).-..))))(((.-...)))....).)))).)))).........- ( -17.30, z-score = -0.06, R) >consensus ____________UGAAAAGAAGAU__UAAUGACAAACAGCAAAUUGCGCUCAAUG_UCGCAUUAAUUGAGGCAAUGA_GCGCAUGAAU_AAAUUCAUGAAUUUACACACCA_AC__A___ .............................................(((((((.((.((.((.....)).)))).))).))))...................................... ( -9.38 = -9.44 + 0.05)
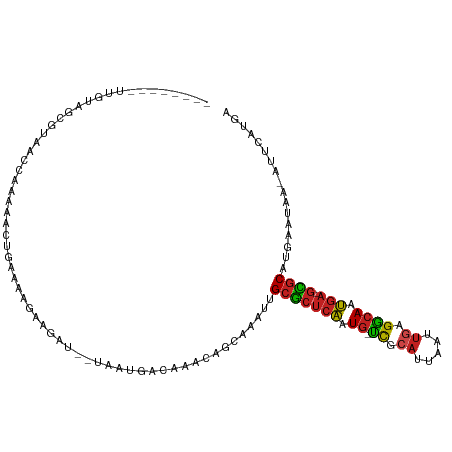
| Location | 14,805,639 – 14,805,740 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Shannon entropy | 0.56471 |
| G+C content | 0.36392 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14805639 101 - 21146708 --------UUGUAGCGUAACCAAAAACUGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGA --------.....((...........((....)).....--...((.....)))).....(((((((.((-((.((.....)).)))).)))-))))(((((...-.))))).. ( -23.10, z-score = -1.68, R) >droSim1.chr2R 13504277 101 - 19596830 --------UUGUAGCGUAACCAAAAACUGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGA --------.....((...........((....)).....--...((.....)))).....(((((((.((-((.((.....)).)))).)))-))))(((((...-.))))).. ( -23.10, z-score = -1.68, R) >droSec1.super_1 12311755 101 - 14215200 --------UUGUAGCGUAACCAAAAACUGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGA --------.....((...........((....)).....--...((.....)))).....(((((((.((-((.((.....)).)))).)))-))))(((((...-.))))).. ( -23.10, z-score = -1.68, R) >droYak2.chr2R 6749057 101 + 21139217 --------UUGUAGCGUAACCAAAAACUGAAAAGAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGA --------.....((...........((....)).....--...((.....)))).....(((((((.((-((.((.....)).)))).)))-))))(((((...-.))))).. ( -23.10, z-score = -1.68, R) >droEre2.scaffold_4845 8997253 101 - 22589142 --------UUGUAGCGUAACCAAAAACUGAAAAUAAGAU--UAAUGACAAACAGCAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUUCAUGA --------...............................--..((((.......((...((((((((.((-((.((.....)).)))).)))-))))))).....-..)))).. ( -21.54, z-score = -1.19, R) >droAna3.scaffold_13266 2351563 107 + 19884421 AAAACCAAAAACCAAGUCUUGCGGCUUGAAA-GGAAGAU--UAAUGAC-AACAGCAAAUUGCGCCCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCAUGAAUAC-AUUCAUGA ............((((((....))))))...-.......--.......-...........((((.((.((-((.((.....)).)))).)).-))))(((((...-.))))).. ( -22.80, z-score = -0.46, R) >dp4.chr3 5881307 86 - 19779522 --------------------GUAGAGGGGGGUUGAAGAU--UAAUGACCAACAGAAAUUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCUUGAAUAA-AUACA--- --------------------(((...(..((((......--....))))..)........(((((((.((-((.((.....)).)))).)))-))))........-.))).--- ( -24.60, z-score = -3.06, R) >droPer1.super_2 6082556 86 - 9036312 --------------------GUGGAGGGGGGUUGAAGAU--UAAUGACAAACAGAAAUUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCUUAAAUAA-AUACA--- --------------------(((....(..(((((...)--))))..)............(((((((.((-((.((.....)).)))).)))-))))........-.))).--- ( -20.60, z-score = -2.18, R) >droWil1.scaffold_180697 2383065 89 - 4168966 ---------------------AAAAAUGCUUAAGAAGAU--UAAUGACAAACAACAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGAGGUGCUUGAAUAA-AUGCAUGA ---------------------....((((((((.....)--)))...(((.((....(((((.((((((.-........)))))))))))....)).))).....-..)))).. ( -17.20, z-score = -0.45, R) >droMoj3.scaffold_6496 20104304 101 + 26866924 ---------UUUGGCCAAAUCACAAAUUGCAAAGAAGAU--UAAUGACAACUGACAAAUUGCGCUCAAUG-UCGCAUUAAUUGAGGCAAUGA-GCGCACAUAUAAGAUGCAUGA ---------..............................--..(((.((.((.......((((((((.((-((.((.....)).)))).)))-)))))......)).))))).. ( -22.82, z-score = -0.87, R) >droGri2.scaffold_15245 17877015 101 - 18325388 --------UUUUGGCCAAGUGAAAAUGAAAAAAGAAGAU--UAAUGACAAAUGACAAAUUGCGCUCAAUG-CCGAAUUAAUUGAGGCAAUGA-GCGCAUGAAUAA-AUGCAUUA --------...............................--(((((.((..........((((((((.((-((...........)))).)))-))))).......-.))))))) ( -22.57, z-score = -2.08, R) >anoGam1.chr3R 22651934 91 - 53272125 ---------------------AAAAGGGGAAGAGAAGAUAAAAACAACGUUCCGUUAGUAGCUGUCGGUGUUUUUUUUUGUUUAGAAAACAA-AAACAGGAAUAA-GUUCAAGG ---------------------..((.((((...(..(.......)..).)))).)).....((((...((((((((.......)))))))).-..))))......-........ ( -11.40, z-score = 0.92, R) >consensus ________UUGUAGCGUAACCAAAAACUGAAAAGAAGAU__UAAUGACAAACAGCAAAUUGCGCUCAAUG_UCGCAUUAAUUGAGGCAAUGA_GCGCAUGAAUAA_AUUCAUGA ............................................................(((((((.((.((.((.....)).)))).))).))))................. ( -9.16 = -9.22 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:12 2011