
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,770,802 – 14,770,931 |
| Length | 129 |
| Max. P | 0.812935 |

| Location | 14,770,802 – 14,770,892 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.26773 |
| G+C content | 0.37668 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
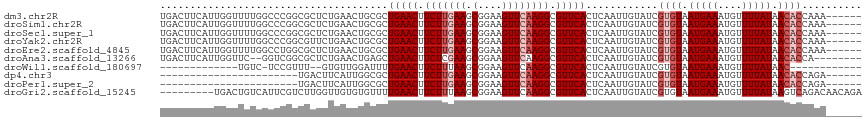
>dm3.chr2R 14770802 90 - 21146708 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAAACAGACACCAAAACAC----------------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).))))....))))............----------------- ( -20.40, z-score = -2.04, R) >droSim1.chr2R 13472020 90 - 19596830 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAAACAGACACCAAAACAC----------------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).))))....))))............----------------- ( -20.40, z-score = -2.04, R) >droSec1.super_1 12277077 90 - 14215200 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAAACAGACACCAAAACAC----------------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).))))....))))............----------------- ( -20.40, z-score = -2.04, R) >droYak2.chr2R 6713521 90 + 21139217 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAAACAGACACCAAAACAC----------------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).))))....))))............----------------- ( -20.40, z-score = -2.04, R) >droEre2.scaffold_4845 8962730 89 - 22589142 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAAACAGACACC-AAACAC----------------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).))))....)))).....-......----------------- ( -20.40, z-score = -1.93, R) >droAna3.scaffold_13266 2312762 85 + 19884421 CUGAACUUCUCGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACAC-----CAGAUACCAAAACAG----------------- .(((((.(((.(((.(....)))).))).))))).......(((((((((.(((((....))))).))))-----..)))))........----------------- ( -19.20, z-score = -1.95, R) >droWil1.scaffold_180697 2336292 77 - 4168966 UUGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACA------CAGACAUC------------------------ ((((((((((......))))))))))..............(((...((((.(((((....))))).)))------)..)))..------------------------ ( -17.50, z-score = -2.03, R) >dp4.chr3 5847016 83 - 19779522 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACA-----CCAGACAACAGAUA------------------- .(((((.(((((((.(....)))))))).))))).((..((((...((((.(((((....))))).)))-----)...))))..))..------------------- ( -22.70, z-score = -2.83, R) >droPer1.super_2 6047175 92 - 9036312 CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACA-----CCAGACAACAGACAACACUGAAC---------- .(((((.(((((((.(....)))))))).))))).....((((...((((.(((((....))))).)))-----)...))))(((......)))...---------- ( -23.70, z-score = -2.25, R) >droGri2.scaffold_15245 17836967 100 - 18325388 -UGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAAGU------CAGACAACAGACAACAAGGAACAGACAACAAG -(((((((((......)))))))))....((((.((...((((....(((..(((..(((...)))..)------)).)))....))))..)))))).......... ( -19.50, z-score = -0.89, R) >consensus CUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACAC____ACAGACACCAAAACAC_________________ .(((((.(((((((.(....)))))))).))))).....(((((.(((........)))...)))))........................................ (-14.53 = -14.83 + 0.30)

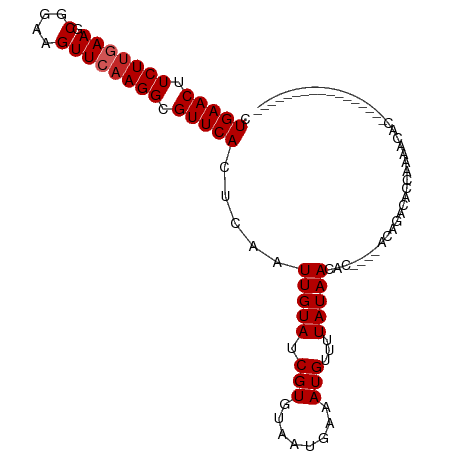

| Location | 14,770,818 – 14,770,929 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.43190 |
| G+C content | 0.41088 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14770818 111 + 21146708 ------UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCA ------....((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...))). ( -28.49, z-score = -1.66, R) >droSim1.chr2R 13472036 111 + 19596830 ------UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCA ------....((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...))). ( -28.49, z-score = -1.66, R) >droSec1.super_1 12277093 111 + 14215200 ------UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCA ------....((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...))). ( -28.49, z-score = -1.66, R) >droYak2.chr2R 6713537 111 - 21139217 ------UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAACGCCGGGCCAAAACCAAUGAAGUCA ------...((((((.........((((..............))))(((((((.(((((((((........))))))))).)).))))).))))))((.......)).......... ( -27.44, z-score = -1.62, R) >droEre2.scaffold_4845 8962745 111 + 22589142 ------UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCAGGCCAAAACCAAUGAAGUCA ------....((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...))). ( -28.49, z-score = -2.14, R) >droAna3.scaffold_13266 2312775 107 - 19884421 --------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCGAGAAGUUCAGCUCAGUUCAGAGCGCCGACC--GAACCAAUGAAGUCA --------.((((((..........((.......))...((((((((((((..((((((.......))))))..))))).)))))))...))))))(((.--...........))). ( -24.30, z-score = -0.89, R) >droWil1.scaffold_180697 2336301 89 + 4168966 ------------GUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUAAAGAAGUUCAAAAUCCAACAC--AAACGGA-GACA------------- ------------(((.............((((.((((...)))).)))).....(((((((((........))))))))).....)))..--....(..-..).------------- ( -14.50, z-score = -1.80, R) >dp4.chr3 5847025 88 + 19779522 ------UCUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCCAAUGAAGUCA----------------------- ------....(((((....)))))((((((.(((...........)))...((.(((((((((........))))))))).)).))))))....----------------------- ( -19.20, z-score = -1.37, R) >droPer1.super_2 6047193 88 + 9036312 ------UCUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCCAAUGAAGUCA----------------------- ------....(((((....)))))((((((.(((...........)))...((.(((((((((........))))))))).)).))))))....----------------------- ( -19.20, z-score = -1.37, R) >droGri2.scaffold_15245 17836988 108 + 18325388 UCUGUUGUCUGACUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUAAAGAAGUUCAAAACACACAACCAAGACGAAUGACAGUCA--------- ...(((((.((.................((((.((((...)))).)))).....(((((((((........)))))))))..)).)))))...(((........))).--------- ( -20.70, z-score = -1.54, R) >consensus ______UUUGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCA .........((((((......(((((.(((.........))).)))))))))))(((((((((........)))))))))..................................... (-12.98 = -13.42 + 0.44)


| Location | 14,770,818 – 14,770,929 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.43190 |
| G+C content | 0.41088 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -16.37 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14770818 111 - 21146708 UGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA------ .((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))....------ ( -29.40, z-score = -1.16, R) >droSim1.chr2R 13472036 111 - 19596830 UGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA------ .((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))....------ ( -29.40, z-score = -1.16, R) >droSec1.super_1 12277093 111 - 14215200 UGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA------ .((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))....------ ( -29.40, z-score = -1.16, R) >droYak2.chr2R 6713537 111 + 21139217 UGACUUCAUUGGUUUUGGCCCGGCGUUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA------ .((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))....------ ( -27.30, z-score = -0.56, R) >droEre2.scaffold_4845 8962745 111 - 22589142 UGACUUCAUUGGUUUUGGCCUGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA------ .((..(.((((((....(((((((((........)))))((((((((......)))))))).))))....)).)))).)..))((((.(((((....))))).))))....------ ( -30.90, z-score = -1.80, R) >droAna3.scaffold_13266 2312775 107 + 19884421 UGACUUCAUUGGUUC--GGUCGGCGCUCUGAACUGAGCUGAACUUCUCGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA-------- ..........((((.--(((..((((..((((((..(((...........)))...))))))..))))..))).)))).....((((.(((((....))))).))))..-------- ( -28.00, z-score = -0.85, R) >droWil1.scaffold_180697 2336301 89 - 4168966 -------------UGUC-UCCGUUU--GUGUUGGAUUUUGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAAC------------ -------------.(((-((((...--....))))..((((((((((......)))))))))))))..........(((((.(((........)))...))))).------------ ( -17.70, z-score = -0.91, R) >dp4.chr3 5847025 88 - 19779522 -----------------------UGACUUCAUUGGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAGA------ -----------------------.((..(.(((((.(.(((((.(((((((.(....)))))))).))))))))))).)..))((((.(((((....))))).))))....------ ( -22.60, z-score = -1.53, R) >droPer1.super_2 6047193 88 - 9036312 -----------------------UGACUUCAUUGGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAGA------ -----------------------.((..(.(((((.(.(((((.(((((((.(....)))))))).))))))))))).)..))((((.(((((....))))).))))....------ ( -22.60, z-score = -1.53, R) >droGri2.scaffold_15245 17836988 108 - 18325388 ---------UGACUGUCAUUCGUCUUGGUUGUGUGUUUUGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAAGUCAGACAACAGA ---------...((((.....((((((..((..((((((((((((((......))))))))))))))..))..)).(((((.(((........)))...)))))...)))).)))). ( -26.90, z-score = -1.38, R) >consensus UGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCAAA______ ......................................(((((.(((((((.(....)))))))).)))))............((((.(((((....))))).)))).......... (-16.37 = -17.07 + 0.70)
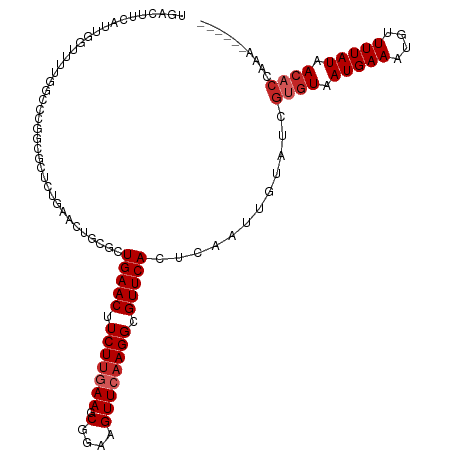

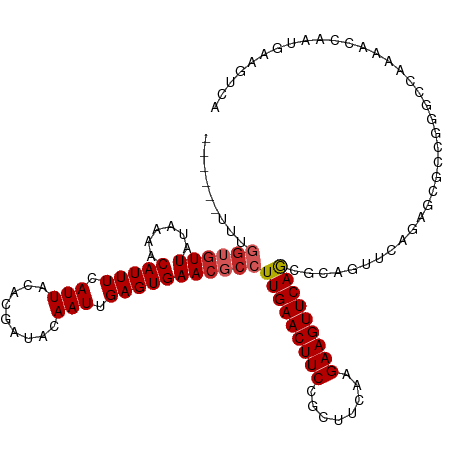
| Location | 14,770,820 – 14,770,931 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.42948 |
| G+C content | 0.41556 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14770820 111 + 21146708 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCAGA ------..((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...)))... ( -28.49, z-score = -1.56, R) >droSim1.chr2R 13472038 111 + 19596830 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCAGA ------..((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...)))... ( -28.49, z-score = -1.56, R) >droSec1.super_1 12277095 111 + 14215200 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCAGA ------..((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...)))... ( -28.49, z-score = -1.56, R) >droYak2.chr2R 6713539 111 - 21139217 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAACGCCGGGCCAAAACCAAUGAAGUCAGA ------.((((((.........((((..............))))(((((((.(((((((((........))))))))).)).))))).))))))((.......))............ ( -27.44, z-score = -1.52, R) >droEre2.scaffold_4845 8962747 111 + 22589142 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCAGGCCAAAACCAAUGAAGUCAGA ------..((((................))))(((...((((.((....((((((((((((........))))))))((((........)))).))))....))))))...)))... ( -28.49, z-score = -2.03, R) >droAna3.scaffold_13266 2312775 109 - 19884421 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCGAGAAGUUCAGCUCAGUUCAGAGCGCCGA--CCGAACCAAUGAAGUCAGA ------.((((((..........((.......))...((((((((((((..((((((.......))))))..))))).)))))))...))))))((--(............)))... ( -24.30, z-score = -0.66, R) >droWil1.scaffold_180697 2336301 91 + 4168966 ----------GUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUAAAGAAGUUCAAAAUCCAACAC--AAACGGA-GACAGA------------- ----------(((.............((((.((((...)))).)))).....(((((((((........))))))))).....)))..--....(..-..)...------------- ( -14.50, z-score = -1.51, R) >dp4.chr3 5847027 88 + 19779522 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCCAAUGAAGUCAGA----------------------- ------..(((((....)))))((((((.(((...........)))...((.(((((((((........))))))))).)).))))))......----------------------- ( -19.20, z-score = -1.28, R) >droPer1.super_2 6047195 88 + 9036312 ------UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCCAAUGAAGUCAGA----------------------- ------..(((((....)))))((((((.(((...........)))...((.(((((((((........))))))))).)).))))))......----------------------- ( -19.20, z-score = -1.28, R) >droGri2.scaffold_15245 17836990 106 + 18325388 UGUUGUCUGACUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUAAAGAAGUUCAAAACACACAACCAAGACGAAUGACAGUCA----------- .(((((.((.................((((.((((...)))).)))).....(((((((((........)))))))))..)).)))))...(((........))).----------- ( -20.70, z-score = -1.73, R) >consensus ______UGGUGUUAUAAAACAUUUCAUUACACGAUACAAUUGAGUGAACGCCUUGAACUUCCGCUUCAAGAAGUUCAGCGCAGUUCAGAGCGCCGGGCCAAAACCAAUGAAGUCAGA .......((((((......(((((.(((.........))).)))))))))))(((((((((........)))))))))....................................... (-12.98 = -13.42 + 0.44)

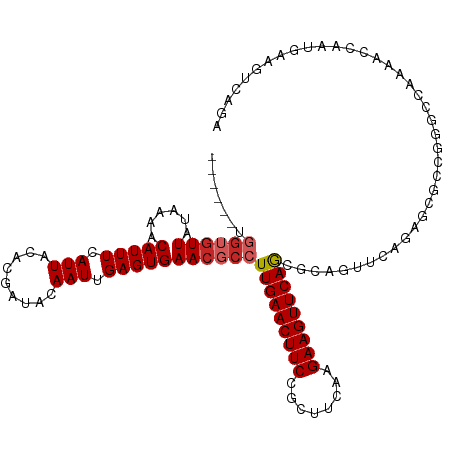

| Location | 14,770,820 – 14,770,931 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.42948 |
| G+C content | 0.41556 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -16.37 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14770820 111 - 21146708 UCUGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ...((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))..------ ( -29.40, z-score = -1.12, R) >droSim1.chr2R 13472038 111 - 19596830 UCUGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ...((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))..------ ( -29.40, z-score = -1.12, R) >droSec1.super_1 12277095 111 - 14215200 UCUGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ...((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))..------ ( -29.40, z-score = -1.12, R) >droYak2.chr2R 6713539 111 + 21139217 UCUGACUUCAUUGGUUUUGGCCCGGCGUUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ...((..(.((((((....(((.(((((........)))))((((((((......))))))))..)))....)).)))).)..))((((.(((((....))))).))))..------ ( -27.30, z-score = -0.52, R) >droEre2.scaffold_4845 8962747 111 - 22589142 UCUGACUUCAUUGGUUUUGGCCUGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ...((..(.((((((....(((((((((........)))))((((((((......)))))))).))))....)).)))).)..))((((.(((((....))))).))))..------ ( -30.90, z-score = -1.75, R) >droAna3.scaffold_13266 2312775 109 + 19884421 UCUGACUUCAUUGGUUCGG--UCGGCGCUCUGAACUGAGCUGAACUUCUCGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ ............((((.((--(..((((..((((((..(((...........)))...))))))..))))..))).)))).....((((.(((((....))))).))))..------ ( -28.00, z-score = -0.76, R) >droWil1.scaffold_180697 2336301 91 - 4168966 -------------UCUGUC-UCCGUUU--GUGUUGGAUUUUGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAAC---------- -------------...(((-((((...--....))))..((((((((((......)))))))))))))..........(((((.(((........)))...))))).---------- ( -17.70, z-score = -0.86, R) >dp4.chr3 5847027 88 - 19779522 -----------------------UCUGACUUCAUUGGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ -----------------------...((..(.(((((.(.(((((.(((((((.(....)))))))).))))))))))).)..))((((.(((((....))))).))))..------ ( -22.60, z-score = -1.84, R) >droPer1.super_2 6047195 88 - 9036312 -----------------------UCUGACUUCAUUGGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA------ -----------------------...((..(.(((((.(.(((((.(((((((.(....)))))))).))))))))))).)..))((((.(((((....))))).))))..------ ( -22.60, z-score = -1.84, R) >droGri2.scaffold_15245 17836990 106 - 18325388 -----------UGACUGUCAUUCGUCUUGGUUGUGUGUUUUGAACUUCUUUAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAAGUCAGACAACA -----------....((((.......(((..((..((((((((((((((......))))))))))))))..))..)))(((((.(((........)))...)))))....))))... ( -26.60, z-score = -1.66, R) >consensus UCUGACUUCAUUGGUUUUGGCCCGGCGCUCUGAACUGCGCUGAACUUCUUGAAGCGGAAGUUCAAGGCGUUCACUCAAUUGUAUCGUGUAAUGAAAUGUUUUAUAACACCA______ ........................................(((((.(((((((.(....)))))))).)))))............((((.(((((....))))).))))........ (-16.37 = -17.07 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:08 2011