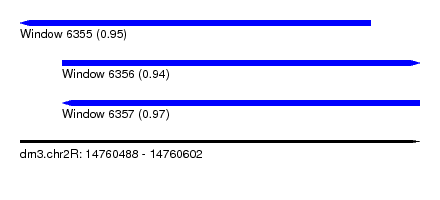
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,760,488 – 14,760,602 |
| Length | 114 |
| Max. P | 0.968134 |

| Location | 14,760,488 – 14,760,588 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Shannon entropy | 0.50824 |
| G+C content | 0.51747 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
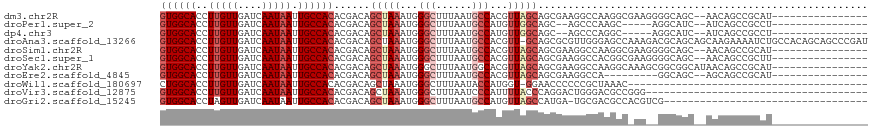
>dm3.chr2R 14760488 100 - 21146708 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGGCGAAGGGGCAGC--AACAGCCGCAU---------------- (((((...((((((..(.....(((((........(((((...(((......)))...))))).((....))...)))))..)..))))--))..)))))..---------------- ( -37.10, z-score = -2.39, R) >droPer1.super_2 6035183 93 - 9036312 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUGGCAGC--AGCCCAAGC-----AGGCAUC--AUCAGCCGCCU---------------- ((((((..(((((....))))).))))))......(((...((((((....((((.....))))..--)))))))))-----.(((...--....)))....---------------- ( -34.20, z-score = -2.14, R) >dp4.chr3 5836443 93 - 19779522 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUGGCAGC--AGCCCAGGC-----AGGCAUC--AUCAGCCGCCU---------------- ((((((..(((((....))))).))))))............((((((....((((.....))))..--))))))(((-----.(((...--....)))))).---------------- ( -36.40, z-score = -2.36, R) >droAna3.scaffold_13266 2300560 117 + 19884421 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUU-GCAGCGCGUUGGGAGCCAAAGACGCAGCAGCAAGAAAAUCUGCCACAGCAGCCCGAU ((((((.(((((((.....................(((.(((((((......))).))))-..)))(((((.(....)...)))))..)))))))......))))))........... ( -35.60, z-score = -0.42, R) >droSim1.chr2R 13461598 100 - 19596830 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGGCGAAGGGGCAGC--AACAGCCGCAU---------------- (((((...((((((..(.....(((((........(((((...(((......)))...))))).((....))...)))))..)..))))--))..)))))..---------------- ( -37.10, z-score = -2.39, R) >droSec1.super_1 12266758 100 - 14215200 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCACGGCGAAGGGGCAGC--AACAGCCGCUU---------------- (((((...((((((..(.....(((((........(((((...(((......)))...))))).((....))...)))))..)..))))--))..)))))..---------------- ( -36.70, z-score = -2.03, R) >droYak2.chr2R 6702985 102 + 21139217 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGGCACGUUAGCAGCGAAGGCCAAGGCAAAGCGGCGGCAUAACAGCCGCAU---------------- ((((((..(((((....))))).))))))......(((((....(((.....)))...))))).((....((....))...)).(((((......)))))..---------------- ( -35.80, z-score = -1.74, R) >droEre2.scaffold_4845 8952430 91 - 22589142 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCA---------GGCAGC--AGCAGCCGCAU---------------- (((((....(((((........(((((........(((((...(((......)))...))))).((....))..---------))))))--)))))))))..---------------- ( -32.30, z-score = -1.80, R) >droWil1.scaffold_180697 2317563 77 - 4168966 CUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUACCAUGGU-GGAACCCCCCGCUAAAC---------------------------------------- .(((((..(((((....))))).)))))......(((...((((.........))))((.-((....)))))))....---------------------------------------- ( -16.70, z-score = -0.46, R) >droVir3.scaffold_12875 859997 81 + 20611582 GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUCCCAUUUUACCCAGGACUGGGACGCCGGG------------------------------------- ((((((..(((((....))))).))))))....(.((.(((((((.......)))))))..((((....))))..)).)..------------------------------------- ( -24.50, z-score = -1.67, R) >droGri2.scaffold_15245 17823632 83 - 18325388 GUGGCACCUAGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUAGCCAUGA-UGCGACGCCACGUCG---------------------------------- ((((((....(((....)))...))))))......(((((...(((......)))...))))).....-..(((((...)))))---------------------------------- ( -24.00, z-score = -1.34, R) >consensus GUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGCC_AA__GGCAGC__AACAGCCGCAU________________ ((((((..(((((....))))).))))))......(((((...(((......)))...)))))....................................................... (-18.18 = -18.97 + 0.79)
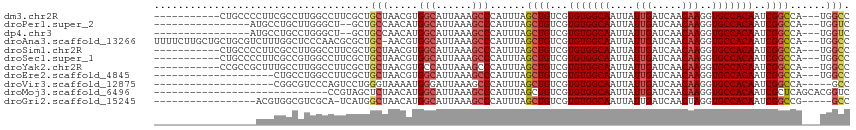
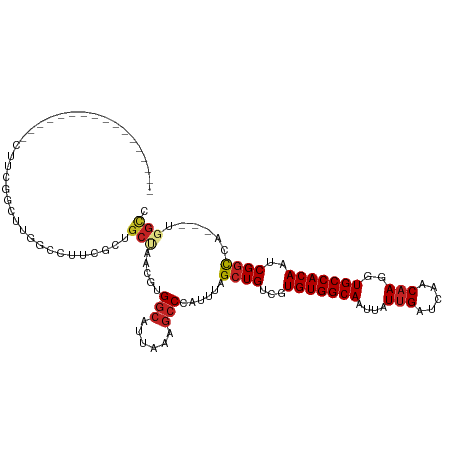
| Location | 14,760,500 – 14,760,602 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Shannon entropy | 0.40253 |
| G+C content | 0.52253 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.944977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14760500 102 + 21146708 -----------CUGCCCCUUCGCCUUGGCCUUCGCUGCUAACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC -----------..........(((.(((((..(((.(((((...(((......)))...))))).).))(((((((....(((.....)))..)))))))...)))))---.))). ( -37.80, z-score = -2.65, R) >droPer1.super_2 6035195 95 + 9036312 ----------------AUGCCUGCUUGGGCU--GCUGCCAACAUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGUC ----------------..(((....((((((--..((((.....))))....))))))....((((...(((((((....(((.....)))..)))))))..))))..---.))). ( -36.60, z-score = -2.11, R) >dp4.chr3 5836455 95 + 19779522 ----------------AUGCCUGCCUGGGCU--GCUGCCAACAUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGUC ----------------......(((((((((--..((((.....))))....))))))....((((...(((((((....(((.....)))..)))))))..))))..---.))). ( -36.50, z-score = -1.93, R) >droAna3.scaffold_13266 2300579 112 - 19884421 UUUUCUUGCUGCUGCGUCUUUGGCUCCCAACGCGCUGC-AACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC .....((((.((.((((...(((...))))))))).))-))((((((.....(((.......)))....(((((((....(((.....)))..)))))))....))))---))... ( -35.20, z-score = -0.49, R) >droSim1.chr2R 13461610 102 + 19596830 -----------CUGCCCCUUCGCCUUGGCCUUCGCUGCUAACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC -----------..........(((.(((((..(((.(((((...(((......)))...))))).).))(((((((....(((.....)))..)))))))...)))))---.))). ( -37.80, z-score = -2.65, R) >droSec1.super_1 12266770 102 + 14215200 -----------CUGCCCCUUCGCCGUGGCCUUCGCUGCUAACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC -----------..........(((((((((..(((.(((((...(((......)))...))))).).))(((((((....(((.....)))..)))))))...)))))---)))). ( -41.70, z-score = -3.60, R) >droYak2.chr2R 6702999 102 - 21139217 -----------CCGCCGCUUUGCCUUGGCCUUCGCUGCUAACGUGCCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC -----------..(((((((((...((((...((.......)).)))).)))))).......((((...(((((((....(((.....)))..)))))))..))))..---.))). ( -33.50, z-score = -1.20, R) >droEre2.scaffold_4845 8952442 93 + 22589142 --------------------CUGCCUGGCCUUCGCUGCUAACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA---UGGCC --------------------..((((((((..(((.(((((...(((......)))...))))).).))(((((((....(((.....)))..)))))))...)))))---.))). ( -36.90, z-score = -2.95, R) >droVir3.scaffold_12875 859999 91 - 20611582 --------------------CGGCGUCCCAGUCCUGGGUAAAAUGGGAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA-----GCC --------------------.(((..((((....))))..(((((((.......))))))).((((...(((((((....(((.....)))..)))))))..))))..-----))) ( -34.60, z-score = -3.05, R) >droMoj3.scaffold_6496 20028937 87 - 26866924 -----------------------------CCGUAGCUCUAACAUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGCUCAGCACGGUC -----------------------------((((...........(((......)))......((((.(((((((((....(((.....)))..)))))))..))..)))))))).. ( -26.80, z-score = -2.39, R) >droGri2.scaffold_15245 17823634 93 + 18325388 -----------------ACGUGGCGUCGCA-UCAUGGCUAACAUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACUAGGUGCCACAAUCGGCCG-----GCC -----------------..(((((((((((-(.((((((((...(((......)))...)))))))).)))))))........(((.....))))))))....((...-----.)) ( -32.60, z-score = -1.63, R) >consensus _________________CUUCGGCUUGGCCUUCGCUGCUAACGUGGCAUUAAAGCCCAUUUAGCUGUCGUGUGGCAAUUAUUGAUCAACAAGGUGCCACAAUCGGCCA___UGGCC ....................................(((.....)))......(((......((((...(((((((....(((.....)))..)))))))..))))......))). (-20.26 = -20.66 + 0.41)
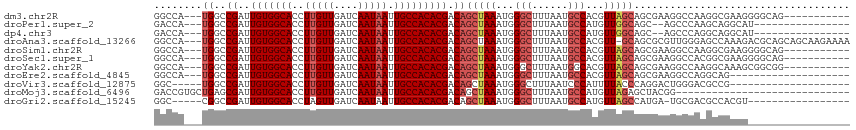
| Location | 14,760,500 – 14,760,602 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Shannon entropy | 0.40253 |
| G+C content | 0.52253 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
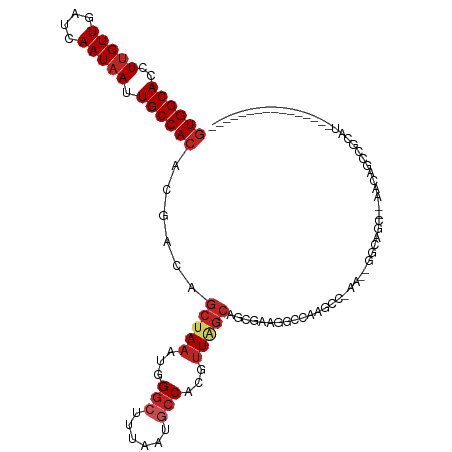
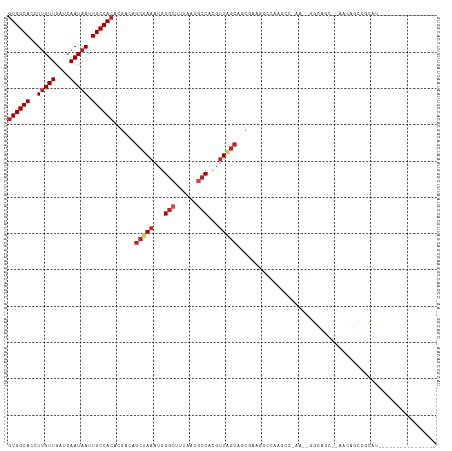
>dm3.chr2R 14760500 102 - 21146708 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGGCGAAGGGGCAG----------- .(((.---(((((...(((((((..(((((....))))).)))))))((...(((((...(((......)))...)))))..))..))))).)))..........----------- ( -41.50, z-score = -3.03, R) >droPer1.super_2 6035195 95 - 9036312 GACCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUGGCAGC--AGCCCAAGCAGGCAU---------------- .....---..(((...(((((((..(((((....))))).))))))).....(((...((((((....((((.....))))..--))))))))).)))..---------------- ( -36.30, z-score = -2.44, R) >dp4.chr3 5836455 95 - 19779522 GACCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUGGCAGC--AGCCCAGGCAGGCAU---------------- .....---..(((...(((((((..(((((....))))).))))))).....(((...((((((....((((.....))))..--))))))))).)))..---------------- ( -36.30, z-score = -2.05, R) >droAna3.scaffold_13266 2300579 112 + 19884421 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUU-GCAGCGCGUUGGGAGCCAAAGACGCAGCAGCAAGAAAA (((..---..)))...(((((((..(((((....))))).))))))).....(((.(((((((......))).))))-...(((((((.(....)...))))).)))))....... ( -36.80, z-score = -0.69, R) >droSim1.chr2R 13461610 102 - 19596830 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGGCGAAGGGGCAG----------- .(((.---(((((...(((((((..(((((....))))).)))))))((...(((((...(((......)))...)))))..))..))))).)))..........----------- ( -41.50, z-score = -3.03, R) >droSec1.super_1 12266770 102 - 14215200 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCACGGCGAAGGGGCAG----------- .(((.---(((((...(((((((..(((((....))))).)))))))((...(((((...(((......)))...)))))..))..))))).)))..........----------- ( -41.50, z-score = -2.95, R) >droYak2.chr2R 6702999 102 + 21139217 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGGCACGUUAGCAGCGAAGGCCAAGGCAAAGCGGCGG----------- .(((.---(((((...(((((((..(((((....))))).)))))))((...(((((....(((.....)))...)))))..))..))))).)))..........----------- ( -38.20, z-score = -1.85, R) >droEre2.scaffold_4845 8952442 93 - 22589142 GGCCA---UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAGGCAG-------------------- .(((.---(((((...(((((((..(((((....))))).)))))))((...(((((...(((......)))...)))))..))..))))))))..-------------------- ( -40.90, z-score = -4.21, R) >droVir3.scaffold_12875 859999 91 + 20611582 GGC-----UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUCCCAUUUUACCCAGGACUGGGACGCCG-------------------- (((-----((..((..(((((((..(((((....))))).))))))))).)))))(((((((.......)))))))..((((....))))......-------------------- ( -31.60, z-score = -2.77, R) >droMoj3.scaffold_6496 20028937 87 + 26866924 GACCGUGCUGAGCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUAGAGCUACGG----------------------------- ..((((((((..((..(((((((..(((((....))))).))))))))).))))......(((((((((.....)))))))))))))----------------------------- ( -30.60, z-score = -3.23, R) >droGri2.scaffold_15245 17823634 93 - 18325388 GGC-----CGGCCGAUUGUGGCACCUAGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCAUGUUAGCCAUGA-UGCGACGCCACGU----------------- (((-----..((((..(((((((....(((....)))...)))))))))...(((((...(((......)))...))))).....-.))...)))....----------------- ( -26.70, z-score = -0.22, R) >consensus GGCCA___UGGCCGAUUGUGGCACCUUGUUGAUCAAUAAUUGCCACACGACAGCUAAAUGGGCUUUAAUGCCACGUUAGCAGCGAAGGCCAAGCCGAAG_________________ ............((..(((((((..(((((....))))).)))))))))...(((((...(((......)))...))))).................................... (-20.85 = -21.26 + 0.41)
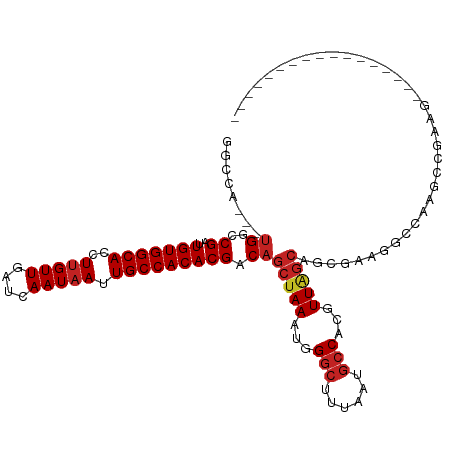
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:33:01 2011