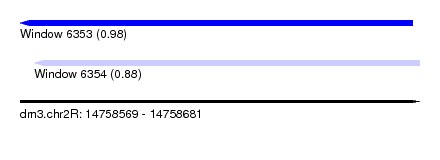
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,758,569 – 14,758,681 |
| Length | 112 |
| Max. P | 0.976936 |

| Location | 14,758,569 – 14,758,679 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Shannon entropy | 0.45695 |
| G+C content | 0.39340 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
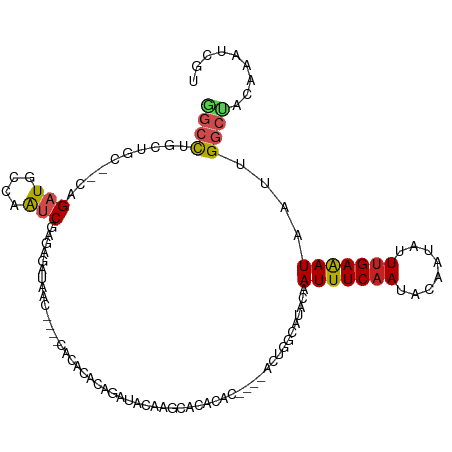
>dm3.chr2R 14758569 110 - 21146708 ---CCUGCUGCAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGUCAUU ---..((((((....))..(((....)))...----..............))))...........(((((....(((((((........)))))))....)))))............ ( -20.30, z-score = -2.23, R) >droAna3.scaffold_13266 2298520 88 + 19884421 ---CCUGUUGGAGAUGCCAAUCGGGAGAUA------------------CAGACACACAC-------AGC-CACAAUUUCAAUAGAAUAUUCGAGAUAAUUGGCUACAAAUCGUCAUU ---((((((((.....)))).)))).....------------------..(((......-------(((-((..(((((((((...)))).)))))...))))).......)))... ( -18.82, z-score = -1.73, R) >droEre2.scaffold_4845 8950572 110 - 22589142 ---CCUGCUGCAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGUCAUU ---..((((((....))..(((....)))...----..............))))...........(((((....(((((((........)))))))....)))))............ ( -20.30, z-score = -2.23, R) >droYak2.chr2R 6700964 117 + 21139217 CCUGCUGCUGCAGAUGCCAAUCGAGAGAUAACCACACACACACAGAUACUAGCACACACUCAAACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGUCAUU .((((....))))(((.(.(((....)))....................................(((((....(((((((........)))))))....)))))......).))). ( -21.90, z-score = -2.12, R) >droSec1.super_1 12264870 110 - 14215200 ---CCUGCUGCAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGUCAUU ---..((((((....))..(((....)))...----..............))))...........(((((....(((((((........)))))))....)))))............ ( -20.30, z-score = -2.23, R) >droSim1.chr2R 13459700 110 - 19596830 ---CCUGCUGCAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGUCAUU ---..((((((....))..(((....)))...----..............))))...........(((((....(((((((........)))))))....)))))............ ( -20.30, z-score = -2.23, R) >droPer1.super_2 6033713 109 - 9036312 ACUACGAC--CAGAUGCCAAUCGAAAGAUAAC--CACACACACAGAUACGAACACACAC----ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUU ...(((((--((((((((((((....)))...--........(......).........----..))))))...(((((((........)))))))..)))).......)))).... ( -17.71, z-score = -3.44, R) >dp4.chr3 5834990 109 - 19779522 ACUACGAC--CAGAUGCCAAUCGAAAGAUAAC--CACACACACAGAUACGAACACACAC----ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUU ...(((((--((((((((((((....)))...--........(......).........----..))))))...(((((((........)))))))..)))).......)))).... ( -17.71, z-score = -3.44, R) >droVir3.scaffold_12875 857166 88 + 20611582 CCUUGUAC---AGAUGCCUGGCCAGAGAUAGC--------------------CACACA------CUGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUU ..(((((.---((.((..((((........))--------------------))..))------))((((....(((((((........)))))))...)))))))))......... ( -18.50, z-score = -2.06, R) >droMoj3.scaffold_6496 20025249 93 + 26866924 CCUUGUACACAAGAUGCUUGGCUAGAGAUAGC--------------------CACACAC----UCUGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUC ..(((((....(((((..((((((....))))--------------------))..)).----)))((((....(((((((........)))))))...)))))))))......... ( -23.30, z-score = -3.81, R) >droGri2.scaffold_15245 17821964 88 - 18325388 CUACGUGC---AGAUCCCUAUCCACAGAUAGC--------------------AACACA------CGGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUU ...((((.---......(((((....))))).--------------------....))------))((((....(((((((........)))))))...)))).............. ( -17.50, z-score = -3.32, R) >consensus ___CCUGCUGCAGAUGCCAAUCGAGAGAUAAC____CACACACAGAUACAAGCACACAC____ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGUCAUU ...............(((.(((....))).............................................(((((((........)))))))....))).............. ( -9.23 = -9.28 + 0.05)

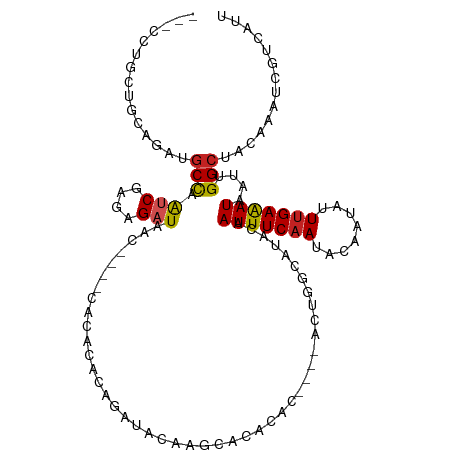
| Location | 14,758,573 – 14,758,681 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Shannon entropy | 0.46195 |
| G+C content | 0.40670 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.44 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
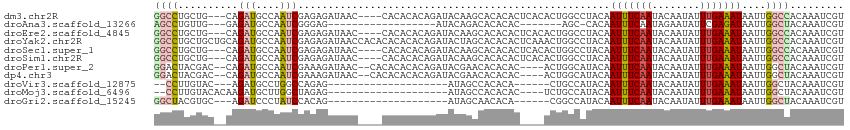
>dm3.chr2R 14758573 108 - 21146708 GGCCUGCUG---CAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGU ((((((...---)))..))).(((....)))...----.............................(((((....(((((((........)))))))....)))))........ ( -20.40, z-score = -1.71, R) >droAna3.scaffold_13266 2298524 86 + 19884421 AGCCUGUUG---GAGAUGCCAAUCGGGAG------------------AUACAGACACACAC-------AGC-CACAAUUUCAAUAGAAUAUUCGAGAUAAUUGGCUACAAAUCGU ..(((((((---(.....)))).))))..------------------..............-------(((-((..(((((((((...)))).)))))...)))))......... ( -17.00, z-score = -1.08, R) >droEre2.scaffold_4845 8950576 108 - 22589142 GGCCUGCUG---CAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGU ((((((...---)))..))).(((....)))...----.............................(((((....(((((((........)))))))....)))))........ ( -20.40, z-score = -1.71, R) >droYak2.chr2R 6700968 115 + 21139217 GGCCUGCUGCUGCAGAUGCCAAUCGAGAGAUAACCACACACACACAGAUACUAGCACACACUCAAACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGU (((((((....))))..))).(((....)))....................................(((((....(((((((........)))))))....)))))........ ( -25.90, z-score = -3.02, R) >droSec1.super_1 12264874 108 - 14215200 GGCCUGCUG---CAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGU ((((((...---)))..))).(((....)))...----.............................(((((....(((((((........)))))))....)))))........ ( -20.40, z-score = -1.71, R) >droSim1.chr2R 13459704 108 - 19596830 GGCCUGCUG---CAGAUGCCAAUCGAGAGAUAAC----CACACACAGAUACAAGCACACACUCACACUGGCCUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCCACAAAUCGU ((((((...---)))..))).(((....)))...----.............................(((((....(((((((........)))))))....)))))........ ( -20.40, z-score = -1.71, R) >droPer1.super_2 6033717 107 - 9036312 GGACUACGAC--CAGAUGCCAAUCGAAAGAUAAC--CACACACACAGAUACGAACACACAC----ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU .....(((((--((((((((((((....)))...--........(......).........----..))))))...(((((((........)))))))..)))).......)))) ( -17.61, z-score = -3.34, R) >dp4.chr3 5834994 107 - 19779522 GGACUACGAC--CAGAUGCCAAUCGAAAGAUAAC--CACACACACAGAUACGAACACACAC----ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU .....(((((--((((((((((((....)))...--........(......).........----..))))))...(((((((........)))))))..)))).......)))) ( -17.61, z-score = -3.34, R) >droVir3.scaffold_12875 857170 84 + 20611582 --CCUUGUAC---AGAUGCCUGGCCAGAG--------------------AUAGCCACACA------CUGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU --..(((((.---((.((..((((.....--------------------...))))..))------))((((....(((((((........)))))))...)))))))))..... ( -18.50, z-score = -2.27, R) >droMoj3.scaffold_6496 20025253 89 + 26866924 --CCUUGUACACAAGAUGCUUGGCUAGAG--------------------AUAGCCACACAC----UCUGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU --..(((((....(((((..((((((...--------------------.))))))..)).----)))((((....(((((((........)))))))...)))))))))..... ( -23.30, z-score = -4.01, R) >droGri2.scaffold_15245 17821968 86 - 18325388 GGCUACGUGC---AGAUCCCUAUCCACAG--------------------AUAGCAACACA------CGGCCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU .....((((.---......(((((....)--------------------)))).....))------))((((....(((((((........)))))))...)))).......... ( -17.50, z-score = -2.49, R) >consensus GGCCUGCUGC__CAGAUGCCAAUCGAGAGAUAAC____CACACACAGAUACAAGCACACAC____ACUGGCAUACAAUUUCAAUACAAUAUUUGAAAUAAUUGGCUACAAAUCGU ((((..........(((....)))....................................................(((((((........)))))))....))))......... ( -8.16 = -8.44 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:58 2011