| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,757,409 – 14,757,501 |
| Length | 92 |
| Max. P | 0.560394 |

| Location | 14,757,409 – 14,757,501 |
|---|---|
| Length | 92 |
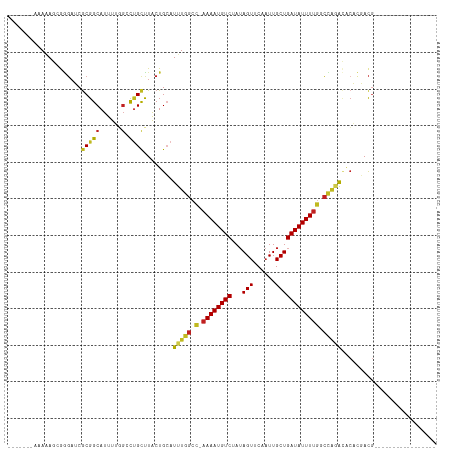
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Shannon entropy | 0.42520 |
| G+C content | 0.44547 |
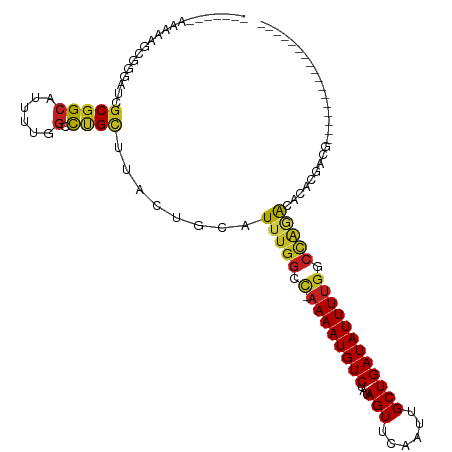
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -10.22 |
| Energy contribution | -10.01 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14757409 92 + 21146708 -------AAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG----------------- -------.....((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.80, R) >droSim1.chr2R 13457942 92 + 19596830 -------AAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCUAGACACACGACG----------------- -------.....((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -30.90, z-score = -3.37, R) >droSec1.super_1 12263718 92 + 14215200 -------AAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG----------------- -------.....((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.80, R) >droYak2.chr2R 6699678 93 - 21139217 ------AAAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG----------------- ------......((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.78, R) >droEre2.scaffold_4845 8949329 93 + 22589142 ------AAAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACC----------------- ------......((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.70, R) >droAna3.scaffold_13266 2297227 99 - 19884421 AAAGAAAAAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG----------------- ............((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.53, R) >dp4.chr3 5833840 90 + 19779522 ---------AAAGCGGGAUCGCGGCAUUUUGGCCUGCCUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG----------------- ---------...((((....(((((......))).))...)))).(((((((-((((((((...(((......)))))))))))))))))).........----------------- ( -33.20, z-score = -3.68, R) >droPer1.super_2 6032556 90 + 9036312 ---------AAAGCGGGAUCGCGGCAUUUUGGCCUGCCUACUGCAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCAAGACACACGACG----------------- ---------...(((....)))(((((((((((((((.....)))...))))-))))))))....(((...((((..(....)..)))))))........----------------- ( -30.40, z-score = -2.95, R) >droWil1.scaffold_180697 2310859 94 + 4168966 ----CCCGAAAAGCAUU-UCGCGGCAUUUUG-UUGGCUUACUGCAUUUGGCCGAAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGAGCCAGCGCCACAUG----------------- ----..(((((....))-))).(((.....(-(((((((.(.((.....)).)((((((((...(((......)))))))))))))))))))))).....----------------- ( -23.70, z-score = -0.50, R) >droVir3.scaffold_12875 855486 101 - 20611582 -----GUACACAAAAAGAGCGCGGCAUUUUCGACUGUUUACUGGAUUUGGGC-AAAAUGUCUAGAGUUCAAUUGCUGAUAUUUUG-CUGUGUGCCACACGAAAAACAC--------- -----(((((((..((((...(((((.........(....)(((((((((((-.....))))))).))))..)))))...)))).-.)))))))..............--------- ( -23.60, z-score = -0.36, R) >droMoj3.scaffold_6496 20023311 112 - 26866924 -AAAAACAAAGAAAAAGAGCGCGGCAUUUGCGGCCGUUUGCUGGAUUUGGCC-AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUGA-CUGUG--GCACACGAAAAAGCCGAAAACACA -................((((((((.......)))))..)))(..((((((.-....(((((((((((.(((.......))).))-)))))--).))).......))))))..)... ( -26.60, z-score = -0.13, R) >droGri2.scaffold_15245 17819592 97 + 18325388 --------GACAAAAAGAGCACGGCAUUUGCGGCUGCUUACUGCAUUUGGCU-AAAAUGUCUAUAGUUCAUUUGCUGAUAUUUUG-CUGAG-GCC-CACGAAAAACACA-------- --------........((((((.((....)).).)))))...((.((..((.-((((((((...(((......))))))))))))-)..))-)).-.............-------- ( -22.00, z-score = -0.03, R) >consensus _______AAAAAGCGGGAUCGCGGCAUUUUGGCCUGCUUACUGCAUUUGGCC_AAAAUGUCUAUAGUUCAAUUGCUGAUAUUUUGGCCAGACACACGACG_________________ ....................(((((......).))))........(((((...((((((((...(((......)))))))))))..))))).......................... (-10.22 = -10.01 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:57 2011