
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,736,491 – 14,736,664 |
| Length | 173 |
| Max. P | 0.774119 |

| Location | 14,736,491 – 14,736,587 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.14 |
| Shannon entropy | 0.69357 |
| G+C content | 0.47151 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -14.07 |
| Energy contribution | -12.95 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.24 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14736491 96 - 21146708 UAUUUCAAGCUCCAACUUACC-------CAUUAUUCUG-----CUAUAGUUUUGUCGAGGGUACUUGCCCGCAUCCUGGUUGCGGCUAUGAAGAUGCCCGCGGUGAUC .......(((...........-------.........)-----))......(..(((.((((((((..(((((.......))))).....))).))))).)))..).. ( -21.05, z-score = 0.76, R) >droSim1.chr2R 13436373 96 - 19596830 UAUUUGAAGCUCAAACUUACC-------UAUUAUUCUG-----UUACAGUUUUGUCGAGGGUACUUGCCCGCAUCCUGGCUGUGGCUAUGAGGAUGCCCGCGGUGAUC ..((((.....)))).(((((-------..((((...(-----((((((((..(..(.((((....)))).)..)..))))))))).))))((....))..))))).. ( -25.00, z-score = -0.29, R) >droSec1.super_1 12244780 96 - 14215200 UAUUUGAAGCUCAAACUUACC-------UAUUAUUCUG-----UUACAGUUUUGUCGAGGGUACUUGCCCGCAUCCUGGCUGUGGCUAUGAGGAUGCCCGCGGUGAUC ..((((.....)))).(((((-------..((((...(-----((((((((..(..(.((((....)))).)..)..))))))))).))))((....))..))))).. ( -25.00, z-score = -0.29, R) >droYak2.chr2R 6680030 103 + 21139217 UUCUUUAAGCUGAACUUUUUUAUUAUUCUGCUAUUUUG-----CUACAGAUUUGUGGAGGGUACUUGCCCGCAUCCCGGCUGUGGCUAUGAGGAUGCCCGUGGUGAUC .......(((.(((...........))).)))...(..-----((((...........((((....))))((((((((((....)))..).))))))..))))..).. ( -26.50, z-score = -0.37, R) >droEre2.scaffold_4845 8928678 96 - 22589142 UGUUUUAAGCUUAAGCUUACU-------UAUUAUUCUG-----CUACAGAUUUGUAGAGGGUACUUGCCCGCAUCCUGGCUGUGGCUAUGAGGAUGCCCGUGGUGAUC .....(((((....))))).(-------((((((((((-----(.........)))))((((....))))((((((((((....)))...)))))))..))))))).. ( -28.70, z-score = -1.22, R) >droAna3.scaffold_13266 2273742 104 + 19884421 -UCUUUUGAUUGAAGAAUUUUGAUUUAUUAUUCUUCUA---AACUUCAGAUUUGUGGAGGGUACUUGCCCGCAUCCUGGUUGCGGCUACGAAGAUGCUCGCGGUGAUC -...(((((..(((((((...........)))))))..---....))))).((((((.((((....))))(((((.(.((.......)).).)))))))))))..... ( -23.90, z-score = -0.12, R) >dp4.chr3 2532556 99 - 19779522 UGCCUUGAAAU-AUAAUCCCUAAAU---UGAGAUUUGC-----CUUCAGCUUUGUGGAGGGCACCUGUCCGCAUCCUGGCUGCGGCUAUGCGGAUGCCCGCGGCGAUC .(((.......-....(((.(((((---((((......-----.))))).)))).)))(((((....(((((((...(((....)))))))))))))))..))).... ( -32.80, z-score = -0.84, R) >droPer1.super_2 2723383 99 - 9036312 UGCCUUGAAAU-AUAAUCCCUAAAU---UGAGAUUUGC-----CUUCAGCUUUGUGGAGGGCACCUGUCCGCAUCCUGGCUGCGGCUAUGCGGAUGCCCGCGGCGAUC .(((.......-....(((.(((((---((((......-----.))))).)))).)))(((((....(((((((...(((....)))))))))))))))..))).... ( -32.80, z-score = -0.84, R) >droWil1.scaffold_180697 2265306 96 - 4168966 ---------UAGAAAAUUCUUGUAAUUAUCUUAUGAUA---UUCAAUAGAUUUGUGGAAGGUACUUGCCCACAUCCAGGCUGCGGCUAUGAAGAUGCACGUGGUGAUC ---------........(((......((((....))))---......)))....((((.(((....)))....)))).((..((((.........)).))..)).... ( -16.70, z-score = 1.29, R) >droVir3.scaffold_12875 832059 102 + 20611582 ---UAUAUGUCUAAAAUUGAAAUGUGUCUAAUUUUCCU---UUUCACAGCUUUGUAGAGGGCACCUGUCCGCAUCCUGGCUGUGGCUAUGAGGACGCACGCGGUGAUC ---............((((...(((((((.....((.(---..((((((((..(....((((....))))....)..))))))))..).)))))))))..)))).... ( -26.60, z-score = -0.48, R) >droMoj3.scaffold_6496 19995661 105 + 26866924 ---UAGAUAUUUAAAAUCUCUUACAUAAUCUACUCUAACCUCCUUUUAGUUUCGUGGAAGGUACCUGUCCACAUCCGGGCUGCGGCUAUGAGGACGCACGCGGCGAUC ---(((((((.(((......))).)).)))))...................(((((((((....)).)))))......((((((((.........)).)))))))).. ( -22.00, z-score = 0.41, R) >droGri2.scaffold_15245 17792937 96 - 18325388 GAAAUUGUAAUCAUAAUUCUCAAUU-------AUAUUU-----CUGUAGCUUUGUGGAGGGCACCUGUCCGCAUCCUGGCUGCGGCUAUGAAGAUGCACGUGGCGAUC (((((.(((((...........)))-------))))))-----)(((((((.((((((.((...)).))))))....)))))))((((((........)))))).... ( -26.60, z-score = -0.87, R) >consensus UACUUUAAGCUCAAAAUUACUAA_U___UAUUAUUCUG_____CUACAGAUUUGUGGAGGGUACUUGCCCGCAUCCUGGCUGCGGCUAUGAGGAUGCCCGCGGUGAUC ........................................................(.((((....)))).)......((((((((.........)).)))))).... (-14.07 = -12.95 + -1.12)


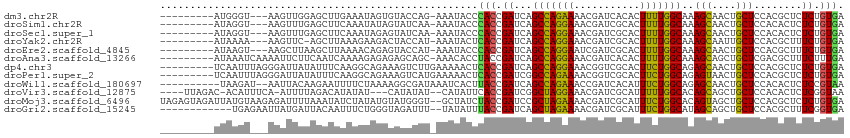
| Location | 14,736,563 – 14,736,664 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.93 |
| Shannon entropy | 0.68182 |
| G+C content | 0.42750 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -11.78 |
| Energy contribution | -10.88 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14736563 101 + 21146708 ---------AUGGGU---AAGUUGGAGCUUGAAAUAGUGUACCAG-AAAUACCCACCGAUCAGCCAGAAAACGAUCACACUUUUGGCAAAGCAACUGCUCCACGCUCUCUGUGA ---------.(((((---(..((((.(((......)))...))))-...)))))).......(((((((...........)))))))..(((....)))...(((.....))). ( -24.50, z-score = -0.55, R) >droSim1.chr2R 13436445 101 + 19596830 ---------AUAGGU---AAGUUUGAGCUUCAAAUAUAGUAUCAA-AAAUACCCACCGAUCAGCCAGGAAACGAUCGCACUUUUGGCAAAGCAACUGCUCCACACUCUCUGUGA ---------...(((---(..((((((((........))).))))-)..))))(((.((...(((((((...........)))))))..(((....)))........)).))). ( -21.10, z-score = -0.91, R) >droSec1.super_1 12244852 101 + 14215200 ---------AUAGGU---AAGUUUGAGCUUCAAAUAGAGUAUCAA-AAAUACCCACCGAUCAGCCAGGAAACGAUCGCACUUUUGGCAAAGCAACUGCUCCACACUCUCUGUGA ---------...(((---(..(((((((((......)))).))))-)..))))(((.((...(((((((...........)))))))..(((....)))........)).))). ( -22.00, z-score = -0.84, R) >droYak2.chr2R 6680110 100 - 21139217 ---------AUAAAA---AAGUUC-AGCUUAAAGAAGACUACCAU-AAAUACUCACCGAUCAGCCAGGAAACGAUCGCACUUUUGGCAAAGCAAUUGCUCCACGCUUUCUGUGA ---------......---......-.(((.((((..((.((....-...)).))..(((((.....(....))))))..)))).)))..(((....)))...(((.....))). ( -13.00, z-score = 1.29, R) >droEre2.scaffold_4845 8928750 101 + 22589142 ---------AUAAGU---AAGCUUAAGCUUAAAACAGAGUACCAU-AAAUACCCACCGAUCAGCCAGGAAUCGAUCGCACUUUUGGCAAAGCAACUGCUCCACGCUUUCUGUGA ---------.....(---((((....)))))..((((((..(...-................(((((((...........)))))))..(((....)))....)..)))))).. ( -18.50, z-score = -0.13, R) >droAna3.scaffold_13266 2273819 104 - 19884421 ---------AUAAAUCAAAAUUCUUCAAUCAAAAGAGAGAGCAGC-AAACACCUACCGAUCAGCCAGGAAACGAUCACACUUUUGGCAAAGCAGCUGCUCGACGCUUUCUUUGA ---------...................(((((((((.(((((((-......(....)....(((((((...........)))))))......)))))))....)))).))))) ( -23.10, z-score = -1.55, R) >dp4.chr3 2532628 105 + 19779522 ---------UCAAUUUAGGGAUUAUAUUUCAAGGCAGAAAGUCUUGAAAAACUCACCGAUCAGCCAGGAAACGGUCGCACUUCUGGCAGAGCAACUGCUCCACGCUCUCUGUGA ---------......((((((.....(((((((((.....))))))))).......((....(((((((...........))))))).((((....))))..)).))))))... ( -29.50, z-score = -1.48, R) >droPer1.super_2 2723455 105 + 9036312 ---------UCAAUUUAGGGAUUAUAUUUCAAGGCAGAAAGUCAUGAAAAACUCACCGAUCGGCCAGAAAACGGUCGCACUUCUGGCAGAGUAACUGCUCCACGCUCUCUGUGA ---------......((((((.....(((((.(((.....))).)))))...........(((((((((...........))))))).((((....))))..)).))))))... ( -23.50, z-score = 0.09, R) >droWil1.scaffold_180697 2265378 102 + 4168966 ----------UAAGAU--AAUUACAAGAAUUUUCUAAAAGGCGAUAAAUCACUUACCGAUCAGCCAGAAACCGAUCACAUUUCUGGCAGAGCAACUGCUCCACACUCUCCGUAA ----------..(((.--((((.....)))).)))....((.((..................((((((((.........)))))))).((((....)))).....)).)).... ( -20.40, z-score = -2.48, R) >droVir3.scaffold_12875 832133 103 - 20611582 ----UUAGAC-ACAUUUCA-AUUUUAGACAUAUAU---CAUAUAU--CAUAUUCACCGAUCGGCUAGGAAACGAUCGCAUUUUUGGCACAGCAGCUGCUCCACACUCUCGGUAA ----......-........-...............---.......--.......(((((...((((((((.........))))))))..(((....)))........))))).. ( -13.40, z-score = 0.31, R) >droMoj3.scaffold_6496 19995732 112 - 26866924 UAGAGUAGAUUAUGUAAGAGAUUUUAAAUAUCUAUAUGUAUGGGU--GCUAUCUACCGAUCCGCUAGAAAACGAUCGCAUUUCUGGCACAGUAGCUGCUCCACGCUCUCUGUGA ..((((((....(((...((((.......))))....(.((.(((--(.....)))).)).)((((((((.........)))))))))))....))))))..(((.....))). ( -25.60, z-score = 0.11, R) >droGri2.scaffold_15245 17793011 100 + 18325388 ------------UGAGAAUUAUGAUUACAAUUUCUGGGUAGAUUU--UAUAUUUACCGAUCAGCUAGAAAACGAUCGCAUUUCUGGCAUAGCAGCUGCUCCACGCUUUCGGUGA ------------.(((.....(((((.((.....))(((((((..--...))))))))))))((((((((.........))))))))..........)))..(((.....))). ( -21.20, z-score = -0.13, R) >consensus _________AUAAGU___AAAUUUUAGCUUAAAAUAGAGAACCAU_AAAUACCCACCGAUCAGCCAGGAAACGAUCGCACUUUUGGCAAAGCAACUGCUCCACGCUCUCUGUGA .....................................................(((.((...(((((((...........)))))))..(((....)))........)).))). (-11.78 = -10.88 + -0.91)


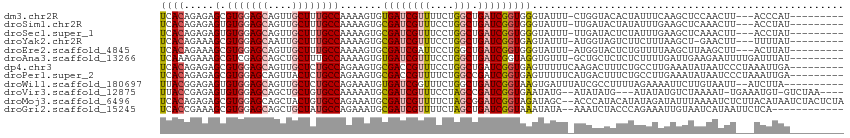
| Location | 14,736,563 – 14,736,664 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.93 |
| Shannon entropy | 0.68182 |
| G+C content | 0.42750 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 14736563 101 - 21146708 UCACAGAGAGCGUGGAGCAGUUGCUUUGCCAAAAGUGUGAUCGUUUUCUGGCUGAUCGGUGGGUAUUU-CUGGUACACUAUUUCAAGCUCCAACUU---ACCCAU--------- ...(((((((((..((((....))))..)((......))...))))))))........(((((((...-.(((....((......))..)))...)---))))))--------- ( -27.60, z-score = -0.45, R) >droSim1.chr2R 13436445 101 - 19596830 UCACAGAGAGUGUGGAGCAGUUGCUUUGCCAAAAGUGCGAUCGUUUCCUGGCUGAUCGGUGGGUAUUU-UUGAUACUAUAUUUGAAGCUCAAACUU---ACCUAU--------- .........(.(..((((....))))..)).......((((((((....))).)))))(((((((..(-((((..((........)).)))))..)---))))))--------- ( -24.70, z-score = -0.19, R) >droSec1.super_1 12244852 101 - 14215200 UCACAGAGAGUGUGGAGCAGUUGCUUUGCCAAAAGUGCGAUCGUUUCCUGGCUGAUCGGUGGGUAUUU-UUGAUACUCUAUUUGAAGCUCAAACUU---ACCUAU--------- .........(.(..((((....))))..)).......((((((((....))).)))))(((((((..(-((((...((.....))...)))))..)---))))))--------- ( -26.40, z-score = -0.52, R) >droYak2.chr2R 6680110 100 + 21139217 UCACAGAAAGCGUGGAGCAAUUGCUUUGCCAAAAGUGCGAUCGUUUCCUGGCUGAUCGGUGAGUAUUU-AUGGUAGUCUUCUUUAAGCU-GAACUU---UUUUAU--------- ....((((..(...((((....))))(((((.((((((.((((..((......)).))))..))))))-.))))))..)))).......-......---......--------- ( -20.40, z-score = 0.93, R) >droEre2.scaffold_4845 8928750 101 - 22589142 UCACAGAAAGCGUGGAGCAGUUGCUUUGCCAAAAGUGCGAUCGAUUCCUGGCUGAUCGGUGGGUAUUU-AUGGUACUCUGUUUUAAGCUUAAGCUU---ACUUAU--------- ..(((((..(((..((((....))))..)((.((((((.(((((((.......))))))).).)))))-.))))..)))))..(((((....))))---).....--------- ( -27.50, z-score = -0.63, R) >droAna3.scaffold_13266 2273819 104 + 19884421 UCAAAGAAAGCGUCGAGCAGCUGCUUUGCCAAAAGUGUGAUCGUUUCCUGGCUGAUCGGUAGGUGUUU-GCUGCUCUCUCUUUUGAUUGAAGAAUUUUGAUUUAU--------- (((((((.((((.((((((.(((((..((((.(((((....)))))..)))).....))))).)))))-).)))).))(((((.....)))))..))))).....--------- ( -28.50, z-score = -0.94, R) >dp4.chr3 2532628 105 - 19779522 UCACAGAGAGCGUGGAGCAGUUGCUCUGCCAGAAGUGCGACCGUUUCCUGGCUGAUCGGUGAGUUUUUCAAGACUUUCUGCCUUGAAAUAUAAUCCCUAAAUUGA--------- ((((.((.(((..(((((.(((((.((......)).))))).).))))..)))..)).))))((.(((((((.(.....).))))))).))..............--------- ( -28.00, z-score = -0.54, R) >droPer1.super_2 2723455 105 - 9036312 UCACAGAGAGCGUGGAGCAGUUACUCUGCCAGAAGUGCGACCGUUUUCUGGCCGAUCGGUGAGUUUUUCAUGACUUUCUGCCUUGAAAUAUAAUCCCUAAAUUGA--------- (((((((((((((((((.(.((((((.((((((((.((....)))))))))).))...)))).).))))))).)))))))...)))...................--------- ( -31.50, z-score = -2.05, R) >droWil1.scaffold_180697 2265378 102 - 4168966 UUACGGAGAGUGUGGAGCAGUUGCUCUGCCAGAAAUGUGAUCGGUUUCUGGCUGAUCGGUAAGUGAUUUAUCGCCUUUUAGAAAAUUCUUGUAAUU--AUCUUA---------- (((((.(((((..(((((....)))))(((((((((.......)))))))))....(((((((...)))))))...........))))))))))..--......---------- ( -26.50, z-score = -1.15, R) >droVir3.scaffold_12875 832133 103 + 20611582 UUACCGAGAGUGUGGAGCAGCUGCUGUGCCAAAAAUGCGAUCGUUUCCUAGCCGAUCGGUGAAUAUG--AUAUAUG---AUAUAUGUCUAAAAU-UGAAAUGU-GUCUAA---- ((((((......(((.((((...)))).)))......)(((((.........)))))))))).....--......(---((((((.((......-.)).))))-)))...---- ( -22.80, z-score = -1.01, R) >droMoj3.scaffold_6496 19995732 112 + 26866924 UCACAGAGAGCGUGGAGCAGCUACUGUGCCAGAAAUGCGAUCGUUUUCUAGCGGAUCGGUAGAUAGC--ACCCAUACAUAUAGAUAUUUAAAAUCUCUUACAUAAUCUACUCUA ....((((.(..(((.((..(((((((((.(((((.((....))))))).)))...))))))...))--..)))..)...(((((((.(((......))).)).))))))))). ( -24.40, z-score = -0.28, R) >droGri2.scaffold_15245 17793011 100 - 18325388 UCACCGAAAGCGUGGAGCAGCUGCUAUGCCAGAAAUGCGAUCGUUUUCUAGCUGAUCGGUAAAUAUA--AAAUCUACCCAGAAAUUGUAAUCAUAAUUCUCA------------ (((((....).)))).(((.(((......)))...)))(((((((....))).))))((((.((...--..)).))))..((((((((....)))))).)).------------ ( -19.30, z-score = -0.22, R) >consensus UCACAGAGAGCGUGGAGCAGUUGCUUUGCCAAAAGUGCGAUCGUUUCCUGGCUGAUCGGUGAGUAUUU_AUGGCUCUCUAUUUUAAGCUAAAACUU___ACUUAU_________ ((((.....(.(((((((....)))))))).......((((((((....))).))))))))).................................................... (-13.00 = -13.27 + 0.26)
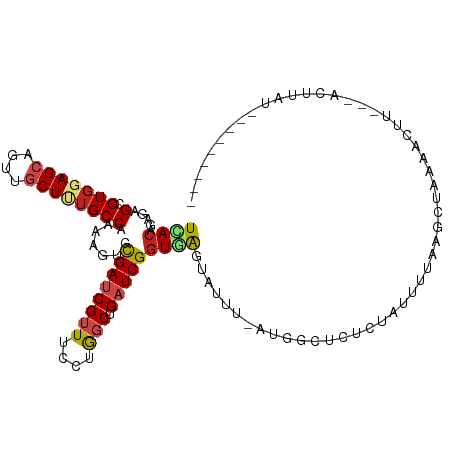
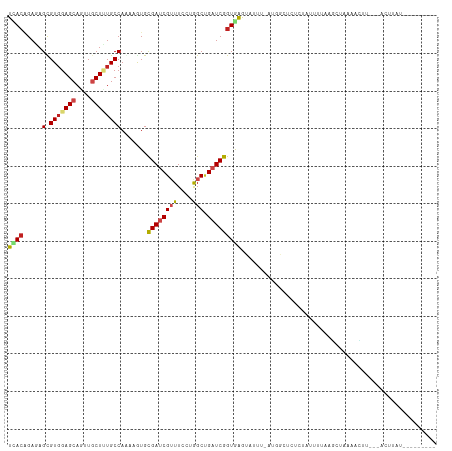
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:55 2011