
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,720,567 – 14,720,682 |
| Length | 115 |
| Max. P | 0.970621 |

| Location | 14,720,567 – 14,720,682 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.62 |
| Shannon entropy | 0.74410 |
| G+C content | 0.52773 |
| Mean single sequence MFE | -34.32 |
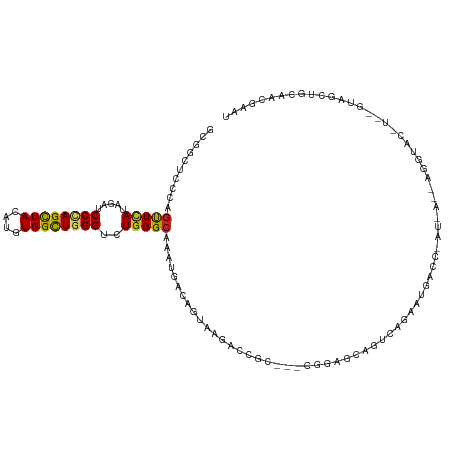
| Consensus MFE | -15.73 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
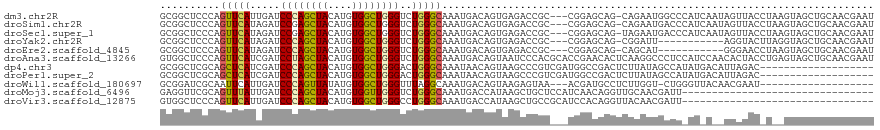
>dm3.chr2R 14720567 115 + 21146708 GCGGCUCCCAGUUCAUUGAUCCCAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUGAGACCGC---CGGAGCAG-CAGAAUGGCCCAUCAAUAGUUACCUAAGUAGCUGCAACGAAU (((((((((((.........((((((((....)))))))).))))((........))))).))))---((..((((-(.((........))..(((....)))....)))))..))... ( -38.20, z-score = -0.76, R) >droSim1.chr2R 13419002 115 + 19596830 GCGGCUCCCAGUUCAUAGAUCCGAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUGAGACCGC---CGGAGCAG-CAGAAUGACCCAUCAAUAGUUACCUAAGUAGCUGCAACGAAU (((((((.(.(((..(((((((.(((((....)))))))))))).......))).).))).))))---((..((((-(....(((....))).(((....)))....)))))..))... ( -36.60, z-score = -1.00, R) >droSec1.super_1 12228671 115 + 14215200 GCGGCUCCCAGUUCAUAGAUCCGAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUGAGACCGC---CGGAGCAG-UAGAAUGACCCAUCAAUAGUUACCUAAGUAGCUGCAACGAAU (((((((.(.(((..(((((((.(((((....)))))))))))).......))).).))).))))---((..((((-(....(((....))).(((....)))....)))))..))... ( -34.30, z-score = -0.53, R) >droYak2.chr2R 6664132 104 - 21139217 GCGGCUCCCAGUUCAUAGAUCCCAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUGAGACCGC---CGGAGCAG-CGGAUU-----------AGGUACUUAGGUAGCUGCAACGAAU ((((((.((.(((((..((.((((((((....))))))))))))))).........(((((((.(---((......-)))...-----------.))).)))))).))))))....... ( -42.40, z-score = -2.52, R) >droEre2.scaffold_4845 8912706 104 + 22589142 GCGGCUCCCAGUUCAUAGAUCCCAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUGAGACCGC---CGGAGCAG-CAGCAU-----------GGGAACCUAAGUAGCUGCAACGAAU (((((((.(.(((..((((.((((((((....)))))))))))).......))).).))).))))---((..((((-(.((.(-----------((....))).)).)))))..))... ( -43.60, z-score = -2.75, R) >droAna3.scaffold_13266 2252872 119 - 19884421 GUGGCUCCCAGUUCAUCGAUCCUAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUAAUCCCACGCACCGAACACUCAAGGCCCUCCAUCCAACACUACCUGAGUAGCUGCAACGAAU (..(((....((((...((.((((((((....))))))))))((((...((....))...))))......))))(((((.((.................))))))))))..)....... ( -32.43, z-score = -0.37, R) >dp4.chr3 2515968 100 + 19779522 GCGGCUCGCAGCUCAUCGAUCCCAGCUACAUGUGGCUGGGACUGGGCAAAUAACAGUAAGCCCGUCGAUGGCCGACUCUUAUAGCCAUAUGACAUUAGAC------------------- ..((((((..((.(((((((((((((((....)))))))))..((((............)))).)))))))))))........)))..............------------------- ( -36.90, z-score = -2.56, R) >droPer1.super_2 2705054 100 + 9036312 GCGGCUCGCAGCUCAUCGAUCCCAGCUACAUGUGGCUGGGACUGGGCAAAUAACAGUAAGCCCGUCGAUGGCCGACUCUUAUAGCCAUAUGACAUUAGAC------------------- ..((((((..((.(((((((((((((((....)))))))))..((((............)))).)))))))))))........)))..............------------------- ( -36.90, z-score = -2.56, R) >droWil1.scaffold_180697 2229335 96 + 4168966 GCGGAUCGCAAUUCAUUGAUCCCAGUUAUAUGUGGCUGGGUUUAGGCAAAUGACAGUAAGAGUAA---ACGAUGCCUCUUGGU-CUGGGUUACAACGAAU------------------- .((((((.........(((.((((((((....)))))))).)))((((....((.......))..---....))))....)))-))).............------------------- ( -23.20, z-score = -0.01, R) >droMoj3.scaffold_6496 19973577 87 - 26866924 GAGGUUCGCAGUUUAUUGAUCCCAGCUACAUGUGGUUGGGUCUGGGCAAAUGACCAUAAGCUGCUCCAUCAACAGGUUGCAACGAUU-------------------------------- ((((...(((((((((.((.((((((((....)))))))))).((........))))))))))).)).)).................-------------------------------- ( -27.60, z-score = -1.35, R) >droVir3.scaffold_12875 7156144 87 - 20611582 GUGGCUCCCAGUUCAUUGAUCCCAGCUACAUGUGGCUGGGCCUGGGCAAAUGACCAUAAGCUGCCGCAUCCACAGGUUACAACGAUU-------------------------------- (((((.(((((.........((((((((....)))))))).)))))................)))))....................-------------------------------- ( -25.43, z-score = 0.22, R) >consensus GCGGCUCCCAGUUCAUAGAUCCCAGCUACAUGUGGCUGGGUCUGGGCAAAUGACAGUAAGACCGC___CGGAGCAGUCAGAAUGACC_AU_A__AGGUAC_U__GUAGCUGCAACGAAU ..........(((((.....((((((((....))))))))..)))))........................................................................ (-15.73 = -15.65 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:52 2011