
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 14,699,802 – 14,699,909 |
| Length | 107 |
| Max. P | 0.500000 |

| Location | 14,699,802 – 14,699,909 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.34 |
| Shannon entropy | 0.46926 |
| G+C content | 0.51072 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -16.91 |
| Energy contribution | -16.44 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 14699802 107 - 21146708 --GCCACUUGACGGCCAUUAUCCCACGAACAGCAACGACACCCGGCUAUCA----AAGGAAACCCUAUCUAGAAUGUCAUCGAGUGGCGCUCGCUAUCU-GGUCAAUGUGAAAC------ --(((((((((.(((.(((.......((..(((..((.....))))).)).----.(((....)))......)))))).)))))))))..((((.....-.......))))...------ ( -24.60, z-score = -0.25, R) >droSim1.chr2R_random 2613583 107 - 2996586 --GCCACUUGACGGCCAUUAUCCCACGAACAGCAACGAGACCCGGCUAUCA----AAGGAAACCCUAUCUAGAAUGUCAUCGAGUGGCGCUCGCUAUCU-GGUCAAUGUGAAAC------ --(((((((((.(((.(((.......((..(((..((.....))))).)).----.(((....)))......)))))).)))))))))..((((.....-.......))))...------ ( -24.60, z-score = -0.01, R) >droSec1.super_1 12208309 85 - 14215200 --GCCACUUGACGGCCAUUAUCCCACGAACA--------------------------GGAAACCCUAUCUAGAAUGUCAUCGAGUGGCGCUCGCUAUCU-GGUCAAUGUGAAAC------ --(((((((((.(((.(((.((....))..(--------------------------((....)))......)))))).)))))))))..((((.....-.......))))...------ ( -21.20, z-score = -0.88, R) >droYak2.chr2R 6642789 111 + 21139217 --GCCACUUGACGGCCAUUAUCCCCCGAACACCAACGACACCGGGCUAUCAUCGAAUGGAAACCCUAUCUGGAAUGUCAUCGAGUGGCGCUCACUAUCU-GGUCAAUGUGGAAG------ --.(((((((((.(((((.....((((..............))))......((((.(((....((.....))....)))))))))))).....(.....-)))))).))))...------ ( -28.14, z-score = 0.10, R) >droEre2.scaffold_4845 8892050 107 - 22589142 --GCCACUUGAAGGCCACUAUCCCCCGAACGCCAAUGACACCGGGCUAUCA----AACGAAACCCUAUCUAGAAUGUCAUCGAGUGGCACUCACUAUCU-GGUCAAUGUGGAAC------ --(((((((((.(((...............)))..(((((..(((...((.----...))..))).........))))))))))))))..((((.....-.......))))...------ ( -24.76, z-score = -0.40, R) >droAna3.scaffold_13266 2229570 99 + 19884421 GAGCCACUUGACAA--ACCA--CCCAGAGC--------CACCCAGU---------GCCGCCAAUUUGACAAAACUGUCAUCAAGUGGCGCUCGCUAUCUCAACAAAAGUGAAACAAAAAG ..(((((((((...--....--....(.((--------(((...))---------)..)))....(((((....))))))))))))))..(((((...........)))))......... ( -23.40, z-score = -2.86, R) >consensus __GCCACUUGACGGCCAUUAUCCCACGAACA_CAACGACACCCGGCUAUCA____AAGGAAACCCUAUCUAGAAUGUCAUCGAGUGGCGCUCGCUAUCU_GGUCAAUGUGAAAC______ ..(((((((((.(((..........................................(....).((....))...))).)))))))))..((((.............))))......... (-16.91 = -16.44 + -0.47)

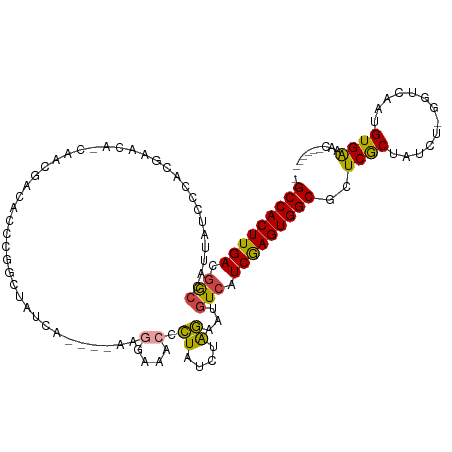
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:32:50 2011